2024-05-26 · Fix template-based method
2024-05-26 · Fix template-based method¶
include("lib/Nto1.jl")
using Revise … ✔ (0.2 s)
using Units, Nto1AdEx, ConnectionTests, ConnTestEval, MemDiskCache … ✔ (0.4 s)
using StatsBase … ✔
N = 6500
duration = 10minutes
600
@time sim = Nto1AdEx.sim(N, duration);
2.564340 seconds (21.50 k allocations: 912.268 MiB, 10.99% gc time)
We won’t test all 6500 inputs (and also not the highest firing), but a random sample.
((_, exc),
(_, inh),
(_, unc)) = inputs = get_trains_to_test(sim, Nₜ = length(sim.trains));
all_trains = [exc; inh; unc]
length(all_trains)
13000
all_trains = [exc; inh; unc]
tvals = []
@showprogress for train in all_trains
push!(tvals, STA_test(sim.V, train.times))
end
(Runtime: 06:51 with 7 threads (and lil interruption). But not threadsafe, adding to a list).
Single threaded: 09:49.
include("lib/plot.jl")
import PythonCall … ✔ (1.2 s)
import PythonPlot … ✔ (2.7 s)
using Sciplotlib … ✔ (0.3 s)
using PhDPlots … ✔
plt.hist(tvals);
θ = quantile(tvals, 0.99)
0.99
sel_trains = all_trains[tvals .≥ θ];
length(sel_trains)
180
STA(train) = calc_STA(sim.V, train.times, ConnectionTests.Δt, 1000);
average(X) = sum(X) / length(X);
template = average([STA(t) for t in sel_trains]);
plotSTA(template);
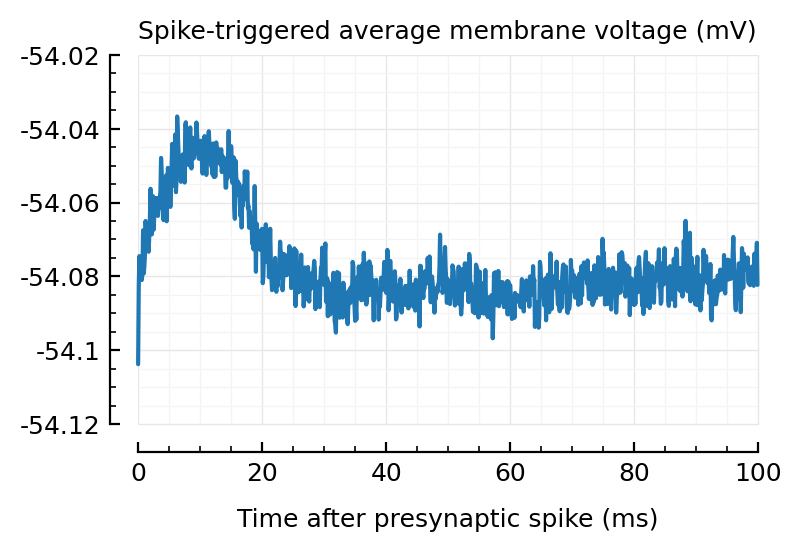
Nice. (We’ll only use the first 20 ms of this).
W = ConnectionTests.STA_length
200
template = template[1:W];
m = TemplateCorr(template);
tvals2 = []
@showprogress for train in all_trains
push!(tvals2, test_conn(m, sim.V, train.times))
end;
Ns = [5, 20, 100, 400, 1600, 6500]
seeds = 1:5;
conntypes(inputs) = begin
conntypes = []
for (conntype, trains) in inputs
append!(conntypes, repeat([conntype], length(trains)))
end
conntypes
end;
rows_template = []
for N in Ns
for seed in seeds
@show N seed
sim = Nto1AdEx.sim(N, duration; seed);
((_, exc),
(_, inh),
(_, unc)) = inputs = get_trains_to_test(sim, Nₜ = length(sim.trains));
all_trains = [exc; inh; unc]
tvals = []
@showprogress "pass1" for train in all_trains
push!(tvals, STA_test(sim.V, train.times))
end
θ = quantile(tvals, 0.99)
sel_trains = all_trains[tvals .≥ θ]
template = average([STA(t) for t in sel_trains])
template = template[1:W]
m = TemplateCorr(template)
tvals2 = []
@showprogress "pass2" for train in all_trains
push!(tvals2, test_conn(m, sim.V, train.times))
end
push!(rows_template, (; N, seed, θ, N_sel=length(sel_trains), tvals, tvals2, conntypes=conntypes(inputs))
end
end;
N = 5
seed = 1
pass1 100%|██████████████████████████████████████████████| Time: 0:00:01
pass2 100%|██████████████████████████████████████████████| Time: 0:00:01
N = 5
seed = 2
pass1 100%|██████████████████████████████████████████████| Time: 0:00:00
pass2 100%|██████████████████████████████████████████████| Time: 0:00:00
N = 5
seed = 3
pass1 100%|██████████████████████████████████████████████| Time: 0:00:01
pass2 100%|██████████████████████████████████████████████| Time: 0:00:01
N = 5
seed = 4
pass1 100%|██████████████████████████████████████████████| Time: 0:00:01
pass2 100%|██████████████████████████████████████████████| Time: 0:00:01
N = 5
seed = 5
pass1 100%|██████████████████████████████████████████████| Time: 0:00:01
pass2 100%|██████████████████████████████████████████████| Time: 0:00:01
N = 20
seed = 1
pass1 100%|██████████████████████████████████████████████| Time: 0:00:02
pass2 100%|██████████████████████████████████████████████| Time: 0:00:02
N = 20
seed = 2
pass1 100%|██████████████████████████████████████████████| Time: 0:00:02
pass2 100%|██████████████████████████████████████████████| Time: 0:00:02
N = 20
seed = 3
pass1 100%|██████████████████████████████████████████████| Time: 0:00:03
pass2 100%|██████████████████████████████████████████████| Time: 0:00:02
N = 20
seed = 4
pass1 100%|██████████████████████████████████████████████| Time: 0:00:01
pass2 100%|██████████████████████████████████████████████| Time: 0:00:02
N = 20
seed = 5
pass1 100%|██████████████████████████████████████████████| Time: 0:00:02
pass2 100%|██████████████████████████████████████████████| Time: 0:00:02
N = 100
seed = 1
pass1 100%|██████████████████████████████████████████████| Time: 0:00:09
pass2 100%|██████████████████████████████████████████████| Time: 0:00:09
N = 100
seed = 2
pass1 100%|██████████████████████████████████████████████| Time: 0:00:08
pass2 100%|██████████████████████████████████████████████| Time: 0:00:08
N = 100
seed = 3
pass1 100%|██████████████████████████████████████████████| Time: 0:00:09
pass2 100%|██████████████████████████████████████████████| Time: 0:00:09
N = 100
seed = 4
pass1 100%|██████████████████████████████████████████████| Time: 0:00:10
pass2 100%|██████████████████████████████████████████████| Time: 0:00:10
N = 100
seed = 5
pass1 100%|██████████████████████████████████████████████| Time: 0:00:12
pass2 100%|██████████████████████████████████████████████| Time: 0:00:11
N = 400
seed = 1
pass1 100%|██████████████████████████████████████████████| Time: 0:00:39
pass2 100%|██████████████████████████████████████████████| Time: 0:00:35
N = 400
seed = 2
pass1 100%|██████████████████████████████████████████████| Time: 0:00:34
pass2 100%|██████████████████████████████████████████████| Time: 0:00:35
N = 400
seed = 3
pass1 100%|██████████████████████████████████████████████| Time: 0:00:35
pass2 100%|██████████████████████████████████████████████| Time: 0:00:35
N = 400
seed = 4
pass1 100%|██████████████████████████████████████████████| Time: 0:00:35
pass2 100%|██████████████████████████████████████████████| Time: 0:00:34
N = 400
seed = 5
pass1 100%|██████████████████████████████████████████████| Time: 0:00:40
pass2 100%|██████████████████████████████████████████████| Time: 0:00:38
N = 1600
seed = 1
pass1 100%|██████████████████████████████████████████████| Time: 0:02:31
pass2 100%|██████████████████████████████████████████████| Time: 0:02:31
N = 1600
seed = 2
pass1 100%|██████████████████████████████████████████████| Time: 0:02:25
pass2 100%|██████████████████████████████████████████████| Time: 0:02:24
N = 1600
seed = 3
pass1 100%|██████████████████████████████████████████████| Time: 0:02:27
pass2 100%|██████████████████████████████████████████████| Time: 0:02:27
N = 1600
seed = 4
pass1 100%|██████████████████████████████████████████████| Time: 0:02:19
pass2 100%|██████████████████████████████████████████████| Time: 0:02:19
N = 1600
seed = 5
pass1 100%|██████████████████████████████████████████████| Time: 0:02:31
pass2 100%|██████████████████████████████████████████████| Time: 0:02:28
N = 6500
seed = 1
pass1 100%|██████████████████████████████████████████████| Time: 0:10:57
pass2 100%|██████████████████████████████████████████████| Time: 0:10:49
N = 6500
seed = 2
pass1 100%|██████████████████████████████████████████████| Time: 0:11:41
pass2 100%|██████████████████████████████████████████████| Time: 0:11:31
N = 6500
seed = 3
pass1 100%|██████████████████████████████████████████████| Time: 0:08:44
pass2 100%|██████████████████████████████████████████████| Time: 0:08:41
N = 6500
seed = 4
pass1 100%|██████████████████████████████████████████████| Time: 0:08:34
pass2 100%|██████████████████████████████████████████████| Time: 0:08:33
N = 6500
seed = 5
pass1 25%|████████████ | ETA: 0:08:50
Total time very approx: 1h10
MemDiskCache.set_dir("2024-05-26__Fix_template-based_method")
"C:\\Users\\tfiers\\.julia\\MemDiskCache.jl\\2024-05-26__Fix_template-based_method"
rows_template_c = @cached rows_template;
Loading [C:\Users\tfiers\.julia\MemDiskCache.jl\2024-05-26__Fix_template-based_method\rows_template.jld2] … ✔ (2.6 s)
perfrows = []
for row in rows_template_c
(; N, seed, tvals2) = row
sweep = sweep_threshold(tvals2, row.conntypes)
AUC = calc_AUROCs(sweep).AUC
F1max = maximum(skipnan(sweep.F1))
push!(perfrows, (; N, seed, method="template-cor", AUC, F1max))
end;
include("lib/df.jl")
using DataFrames … ✔ (0.5 s)
df = DataFrame(perfrows);
gdf = groupby(df, :N)
combine(gdf, nrow => :num_seeds, [:AUC, :F1max] .=> mean)
| Row | N | num_seeds | AUC_mean | F1max_mean |
|---|---|---|---|---|
| Int64 | Int64 | Float64 | Float64 | |
| 1 | 5 | 5 | 0.98 | 0.982 |
| 2 | 20 | 5 | 0.985 | 0.986 |
| 3 | 100 | 5 | 0.975 | 0.976 |
| 4 | 400 | 5 | 0.977 | 0.971 |
| 5 | 1600 | 5 | 0.884 | 0.83 |
| 6 | 6500 | 5 | 0.511 | 0.529 |
:D
We’ll repeat STA and linefit as well.
Actually for STA we already have the tvals!
for row in rows_template_c
(; N, seed, tvals) = row
sweep = sweep_threshold(tvals, row.conntypes)
AUC = calc_AUROCs(sweep).AUC
F1max = maximum(skipnan(sweep.F1))
push!(perfrows, (; N, seed, method="STA", AUC, F1max))
end;
And now:
m = ConnectionTests.FitUpstroke()
FitUpstroke(100, 0)
linefit() = begin
rows = []
for N in Ns
for seed in seeds
@show N seed
sim = Nto1AdEx.sim(N, duration; seed);
((_, exc),
(_, inh),
(_, unc)) = inputs = get_trains_to_test(sim, Nₜ = length(sim.trains));
all_trains = [exc; inh; unc]
tvals = []
@showprogress for train in all_trains
push!(tvals, test_conn(m, sim.V, train.times))
end
push!(rows, (; N, seed, tvals, conntypes = conntypes(inputs)))
end
end
rows
end
rows_linefit = @cached linefit();
Loading [C:\Users\tfiers\.julia\MemDiskCache.jl\2024-05-26__Fix_template-based_method\linefit().jld2] … ✔ (1.9 s)
Approx runtime: 16 minutes.
Vs the 1h10 for two-pass STA / template corr: 4x faster.
Discovering sth new here – that makes sweep_threshold takes a lot of time and memory for linefit:
rows_template_c[end].tvals |> unique |> length
200
rows_linefit[end].tvals |> unique |> length
13000
(Not doing anything about it now)
(Some info already though: PredictionTable should be fully type stable, so efficient memory representation ig.
Next step: how big is one such PredictionTable).
@showprogress for row in rows_linefit
(; N, seed, tvals) = row
sweep = sweep_threshold(tvals, row.conntypes)
AUC = calc_AUROCs(sweep).AUC
F1max = maximum(skipnan(sweep.F1))
push!(perfrows, (; N, seed, method="linefit", AUC, F1max))
end;
Progress: 100%|█████████████████████████████████████████| Time: 0:02:39
perfrows_c = @cached perfrows;
deleteat!(perfrows_c, 61); # duplicate row
Loading [C:\Users\tfiers\.julia\MemDiskCache.jl\2024-05-26__Fix_template-based_method\perfrows.jld2] … ✔ (2.2 s)
include("lib/df.jl")
using DataFrames … ✔
df = DataFrame(perfrows_c)
| Row | N | seed | method | AUC | F1max |
|---|---|---|---|---|---|
| Int64 | Int64 | String | Float64 | Float64 | |
| 1 | 5 | 1 | template-cor | 1 | 1 |
| 2 | 5 | 2 | template-cor | 1 | 1 |
| 3 | 5 | 3 | template-cor | 1 | 1 |
| 4 | 5 | 4 | template-cor | 0.9 | 0.909 |
| 5 | 5 | 5 | template-cor | 1 | 1 |
| ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ |
| 86 | 6500 | 1 | linefit | 0.469 | 0.496 |
| 87 | 6500 | 2 | linefit | 0.495 | 0.515 |
| 88 | 6500 | 3 | linefit | 0.489 | 0.509 |
| 89 | 6500 | 4 | linefit | 0.511 | 0.525 |
| 90 | 6500 | 5 | linefit | 0.481 | 0.504 |
gdf = groupby(df, [:method, :N])
dfm = combine(gdf, nrow => :num_seeds, [:AUC, :F1max] .=> mean)
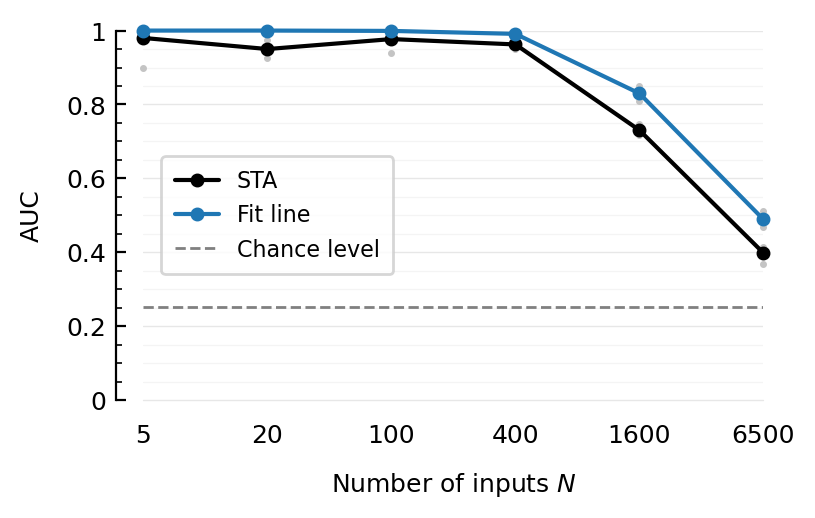
sort(dfm[dfm.N .>= 400, :], :N)
| Row | method | N | num_seeds | AUC_mean | F1max_mean |
|---|---|---|---|---|---|
| String | Int64 | Int64 | Float64 | Float64 | |
| 1 | template-cor | 400 | 5 | 0.977 | 0.971 |
| 2 | STA | 400 | 5 | 0.962 | 0.941 |
| 3 | linefit | 400 | 5 | 0.991 | 0.966 |
| 4 | template-cor | 1600 | 5 | 0.884 | 0.83 |
| 5 | STA | 1600 | 5 | 0.73 | 0.687 |
| 6 | linefit | 1600 | 5 | 0.83 | 0.771 |
| 7 | template-cor | 6500 | 5 | 0.511 | 0.529 |
| 8 | STA | 6500 | 5 | 0.398 | 0.471 |
| 9 | linefit | 6500 | 5 | 0.489 | 0.51 |
Cool. STA-based two-pass template corr even better than linefit.
Now plot.
include("lib/plot.jl")
import PythonCall … ✔
import PythonPlot … ✔
using Sciplotlib … ✔
using PhDPlots … ✔
chance_AUC = 0.252;
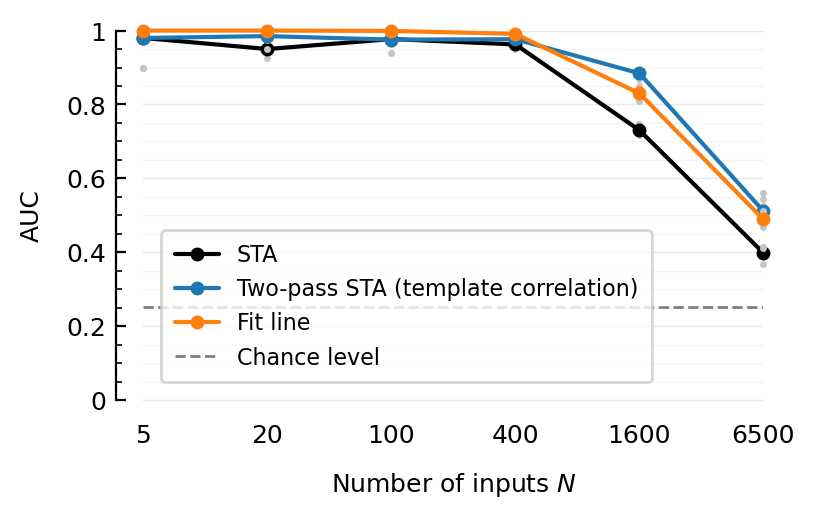
ax = newax()
plotAUC(ax, m; kw...) = begin
plot_dots_and_means(
df[df.method.==m, :], :N, :AUC;
ax,
xtype=:categorical, xticklabels=Ns,
ylim=[0,1],
xlabel = L"Number of inputs $N$",
kw...
)
end
add_chance_line(ax) = ax.axhline(chance_AUC, ls="--", lw=1, color="gray", label="Chance level")
add_chance_line(ax)
plotAUC(ax, "STA", line_label="STA");
plotAUC(ax, "template-cor", color_means=C0, line_label="Two-pass STA (template correlation)");
plotAUC(ax, "linefit", color_means=C1, line_label="Fit line");
legend(ax, reorder=[1=>4]);
ax = newax()
add_chance_line(ax)
plotAUC(ax, "STA", line_label="STA");
plotAUC(ax, "linefit", color_means=C0, line_label="Fit line");
legend(ax, reorder=[1=>3]);
savefig_phd("perf-all-inputs__fitline")
Saved at `../thesis/figs/perf-all-inputs__fitline.pdf`
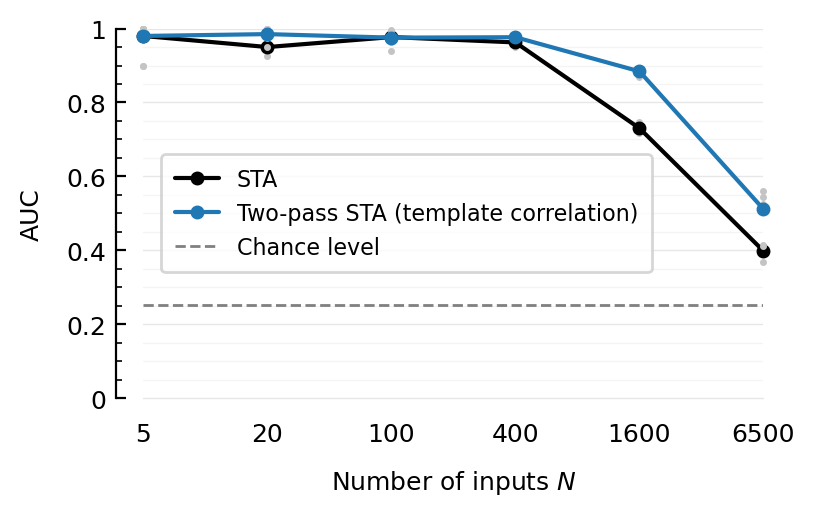
ax = newax()
add_chance_line(ax)
plotAUC(ax, "STA", line_label="STA");
plotAUC(ax, "template-cor", color_means=C0, line_label="Two-pass STA (template correlation)");
legend(ax, reorder=[1=>3]);
savefig_phd("perf-all-inputs__template-cor")
Saved at `../thesis/figs/perf-all-inputs__template-cor.pdf`