2022-03-02 • Duration & SNR for big-N–to–1
Contents
2022-03-02 • Duration & SNR for big-N–to–1¶
Setup¶
#
using Revise
using MyToolbox
using VoltageToMap
[ Info: Precompiling VoltageToMap [b3b8fdc5-3c26-4000-a0c8-f17415fdf48e]
Params & sim¶
Short warm-up run. Get compilation out of the way.
p0 = ExperimentParams(
sim = SimParams(
input = previous_N_30_input,
duration = 1 * minutes
)
);
@time sim(p0.sim);
Progress: 100%|█████████████████████████████████████████| Time: 0:00:01
3.929238 seconds (8.80 M allocations: 598.610 MiB, 11.79% gc time, 86.25% compilation time)
p = ExperimentParams(
sim = SimParams(
input = realistic_N_6600_input,
duration = 0.2 * minutes,
synapses = SynapseParams(
Δg_multiplier = 0.066,
),
)
);
dumps(p)
ExperimentParams
rngseed: 22022022
sim: SimParams
duration: 12.0
Δt: 0.0001
num_timesteps: 120000
rngseed: 0
input: PoissonInputParams
N_unconn: 100
N_exc: 5200
N_inh: 1300
N_conn: 6500
N: 6600
spike_rates: LogNormal
μ: 1.08629
σ: 0.774597
synapses: SynapseParams
Δg_exc: 4.0e-10
Δg_inh: 1.6e-9
Δg_multiplier: 0.066
E_exc: 0.0
E_inh: -0.065
g_t0: 0.0
τ: 0.007
izh_neuron: IzhikevichParams
C: 1.0e-10
k: 7.0e-7
v_rest: -0.06
v_thr: -0.04
a: 30.0
b: -2.0e-9
v_peak: 0.035
v_reset: -0.05
Δu: 1.0e-10
v_t0: -0.06
u_t0: 0.0
imaging: VoltageImagingParams
spike_SNR: 10.0
spike_SNR_dB: 20.0
spike_height: 0.095
σ_noise: 0.0095
conntest: ConnTestParams
STA_window_length: 0.1
num_shuffles: 100
rngseed: 22022022
evaluation: EvaluationParams
num_tested_neurons_per_group: 40
rngseed: 22022022
t, v, vimsig, input_spikes = @time sim(p.sim);
Progress: 100%|█████████████████████████████████████████| Time: 0:00:02
4.708045 seconds (4.80 M allocations: 372.494 MiB, 2.00% gc time, 39.88% compilation time)
num_spikes = length.(input_spikes)
ComponentVector{Int64}(conn = (exc = [24, 18, 27, 22, 12, 23, 20, 6, 18, 18 … 20, 18, 26, 22, 7, 12, 26, 8, 22, 15], inh = [7, 19, 16, 1, 19, 13, 21, 22, 3, 14 … 27, 15, 21, 7, 22, 26, 20, 17, 18, 27]), unconn = [22, 18, 15, 22, 29, 17, 12, 29, 14, 14 … 29, 16, 20, 4, 28, 27, 28, 11, 10, 8])
Plot¶
import PyPlot
using VoltageToMap.Plot
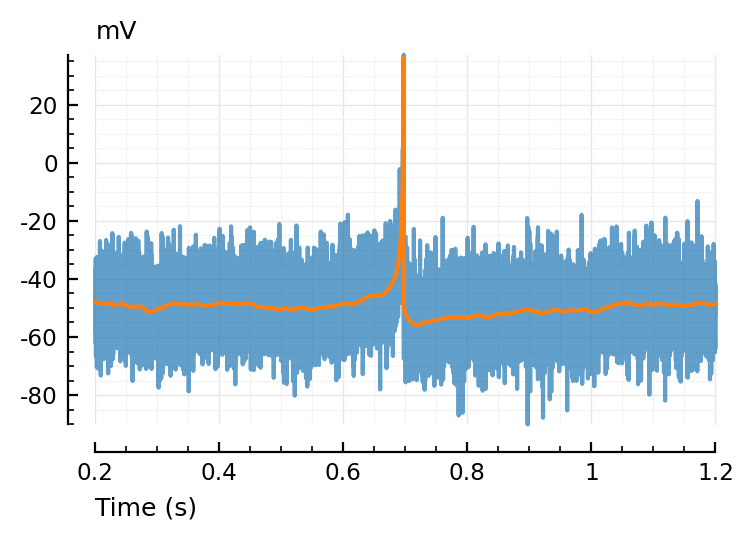
tzoom = [200, 1200]ms
ax = plotsig(t, vimsig / mV, tzoom; xlabel="Time (s)", hylabel="mV", alpha=0.7);
plotsig(t, v / mV, tzoom; ax);
Test conntest¶
example_presynspikes = input_spikes.conn.exc[44]
plotSTA(vimsig, example_presynspikes, p);
p_value = test_connection(vimsig, example_presynspikes, p)
0.61
Conntest performance¶
N_eval_trains = p.evaluation.num_tested_neurons_per_group
40
α = 0.05;
function evaluate_conntest_performance(vimsig, input_spikes, p)
resetrng!(p.evaluation.rngseed)
TP_exc = 0
TP_inh = 0
TP_unconn = 0
for input_train in input_spikes.conn.exc[1:N_eval_trains]
p_value = test_connection(vimsig, input_train, p)
if p_value < α
TP_exc += 1
end
end
for input_train in input_spikes.conn.inh[1:N_eval_trains]
p_value = test_connection(vimsig, input_train, p)
if p_value > 1 - α
TP_inh += 1
end
end
for input_train in input_spikes.unconn[1:N_eval_trains]
p_value = test_connection(vimsig, input_train, p)
if α/2 ≤ p_value ≤ 1 - α/2
TP_unconn += 1
end
end
TPR_exc = TP_exc / N_eval
TPR_inh = TP_inh / N_eval
TPR_unconn = TP_unconn / N_eval
FPR = 1 - TPR_unconn
return TPR_exc, TPR_inh, FPR
end;
evaluate_conntest_performance(vimsig, input_spikes, p)
(0.05, 0.025, 0.025000000000000022)
Performance for given params¶
function performance_for(p::ExperimentParams)
_t, _v, vimsig, input_spikes = sim(p.sim);
return evaluate_conntest_performance(vimsig, input_spikes, p)
end;
VI_params = VoltageImagingParams(;
spike_height = cortical_RS.v_peak - cortical_RS.v_rest,
spike_SNR = Inf,
);
durations = [
30 * seconds,
1 * minutes,
5 * minutes,
10 * minutes,
20 * minutes,
30 * minutes,
]
xlabels = durations / minutes .|> x -> @sprintf "%.3G" x;
TPRs_exc = Vector{Float64}()
TPRs_inh = Vector{Float64}()
FPRs = Vector{Float64}()
for duration in durations
@show duration / minutes
params = ExperimentParams(sim = SimParams(; duration, imaging = VI_params))
TPR_exc, TPR_inh, FPR = performance_for(params)
@show TPR_exc, TPR_inh, FPR
push!(TPRs_exc, TPR_exc)
push!(TPRs_inh, TPR_inh)
push!(FPRs, FPR)
println()
end
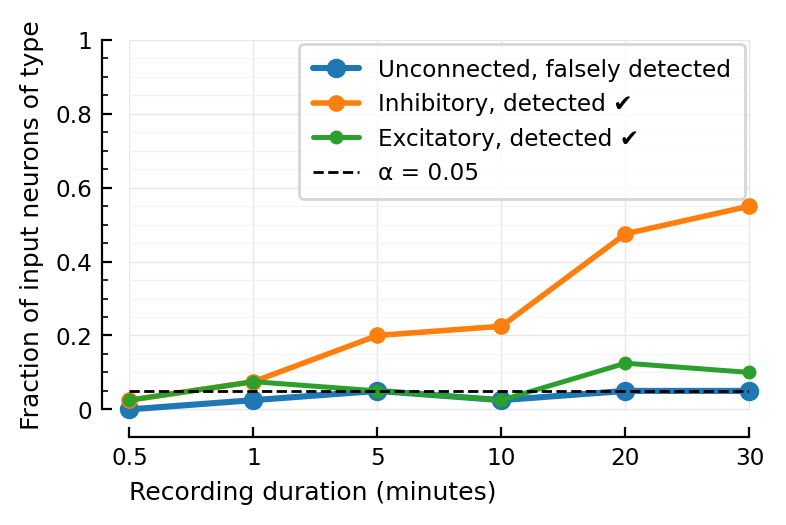
duration / minutes = 0.5
Progress: 100%|█████████████████████████████████████████| Time: 0:00:06
(TPR_exc, TPR_inh, FPR) = (0.025, 0.025, 0.0)
duration / minutes = 1.0
Progress: 100%|█████████████████████████████████████████| Time: 0:00:15
(TPR_exc, TPR_inh, FPR) = (0.075, 0.075, 0.025000000000000022)
duration / minutes = 5.0
Progress: 100%|█████████████████████████████████████████| Time: 0:01:26
(TPR_exc, TPR_inh, FPR) = (0.05, 0.2, 0.050000000000000044)
duration / minutes = 10.0
Progress: 100%|█████████████████████████████████████████| Time: 0:02:30
(TPR_exc, TPR_inh, FPR) = (0.025, 0.225, 0.025000000000000022)
duration / minutes = 20.0
Progress: 100%|█████████████████████████████████████████| Time: 0:05:24
(TPR_exc, TPR_inh, FPR) = (0.125, 0.475, 0.050000000000000044)
duration / minutes = 30.0
Progress: 100%|█████████████████████████████████████████| Time: 0:07:40
(TPR_exc, TPR_inh, FPR) = (0.1, 0.55, 0.050000000000000044)
xticks = [1:length(durations);]
plott(rates; kw...) = plot(
xticks, rates, ".-"; ylim=(0,1),
xminorticks=false, clip_on=false, kw...
)
smaller = (lw=1.8, ms=08)
medium = (lw=2.0, ms=10)
larger = (lw=2.2, ms=12)
ax = plott(FPRs; larger..., label="Unconnected, falsely detected")
ax = plott(TPRs_inh; medium..., label="Inhibitory, detected ✔")
ax = plott(TPRs_exc; smaller..., label="Excitatory, detected ✔")
ax.set_xticks(xticks, xlabels)
ax.set_xlabel("Recording duration (minutes)")
ax.set_ylabel("Fraction of input neurons of type")
ax.axhline(α, color="black", zorder=3, lw=1, linestyle="dashed", label=f"α = {α:.3G}")
ax.legend();