2022-10-24 • N-to-1 with lognormal inputs
Contents
2022-10-24 • N-to-1 with lognormal inputs¶
Imports¶
#
using Revise
@time using MyToolbox
@time using SpikeWorks
@time using Sciplotlib
@time using VoltoMapSim
WARNING: using MyToolbox.@withfb in module Main conflicts with an existing identifier.
6.076024 seconds (3.39 M allocations: 212.208 MiB, 2.41% gc time, 32.60% compilation time: 61% of which was recompilation)
4.639393 seconds (2.65 M allocations: 159.224 MiB, 2.82% gc time, 32.20% compilation time: 86% of which was recompilation)
23.643910 seconds (13.39 M allocations: 754.670 MiB, 3.50% gc time, 56.14% compilation time: 62% of which was recompilation)
13.120539 seconds (9.23 M allocations: 595.361 MiB, 4.11% gc time, 2.81% compilation time: 16% of which was recompilation)
Start¶
Neuron-model parameters¶
@typed begin
# Izhikevich params
C = 100 * pF # Cell capacitance
k = 0.7 * (nS/mV) # Steepness of parabola in v̇(v)
vₗ = - 60 * mV # Resting ('leak') membrane potential
vₜ = - 40 * mV # Spiking threshold (when no syn. & adaptation currents)
a = 0.03 / ms # Reciprocal of time constant of adaptation current `u`
b = - 2 * nS # (v-vₗ)→u coupling strength
vₛ = 35 * mV # Spike cutoff (defines spike time)
vᵣ = - 50 * mV # Reset voltage after spike
Δu = 100 * pA # Adaptation current inflow on self-spike
# Conductance-based synapses
Eₑ = 0 * mV # Reversal potential at excitatory synapses
Eᵢ = -80 * mV # Reversal potential at inhibitory synapses
τ = 7 * ms # Time constant for synaptic conductances' decay
end;
Simulated variables and their initial values¶
x₀ = (
# Izhikevich variables
v = vᵣ, # Membrane potential
u = 0 * pA, # Adaptation current
# Synaptic conductances g
gₑ = 0 * nS, # = Sum over all exc. synapses
gᵢ = 0 * nS, # = Sum over all inh. synapses
);
Differential equations:¶
calculate time derivatives of simulated vars
(and store them “in-place”, in Dₜ).
function f!(Dₜ, vars)
v, u, gₑ, gᵢ = vars
# Conductance-based synaptic current
Iₛ = gₑ*(v-Eₑ) + gᵢ*(v-Eᵢ)
# Izhikevich 2D system
Dₜ.v = (k*(v-vₗ)*(v-vₜ) - u - Iₛ) / C
Dₜ.u = a*(b*(v-vₗ) - u)
# Synaptic conductance decay
Dₜ.gₑ = -gₑ / τ
Dₜ.gᵢ = -gᵢ / τ
end;
Spike discontinuity¶
has_spiked(vars) = (vars.v ≥ vₛ)
function on_self_spike!(vars)
vars.v = vᵣ
vars.u += Δu
end;
Conductance-based Izhikevich neuron¶
coba_izh_neuron = NeuronModel(x₀, f!; has_spiked, on_self_spike!);
More parameters, and input spikers¶
using SpikeWorks.Units
using SpikeWorks: LogNormal
Δt = 0.1ms # Sim timestep
sim_duration = 10seconds
sim_duration = 1minute
sim_duration = 10minutes
600
Firing rates λ for the Poisson inputs
fr_distr = LogNormal(median = 4Hz, g = 2)
Distributions.LogNormal{Float64}(μ=1.39, σ=0.693)
@enum NeuronType exc inh
input(;
N = 100,
EIratio = 4//1,
scaling = N,
) = begin
firing_rates = rand(fr_distr, N)
input_IDs = 1:N
inputs = [
Nto1Input(ID, poisson_SpikeTrain(λ, sim_duration))
for (ID, λ) in zip(input_IDs, firing_rates)
]
# Nₑ, Nᵢ = groupsizes(EIMix(N, EIratio))
EImix = EIMix(N, EIratio)
Nₑ = EImix.Nₑ
Nᵢ = EImix.Nᵢ
neuron_type(ID) = (ID ≤ Nₑ) ? exc : inh
Δgₑ = 60nS / scaling
Δgᵢ = 60nS / scaling * EIratio
on_spike_arrival!(vars, spike) =
if neuron_type(source(spike)) == exc
vars.gₑ += Δgₑ
else
vars.gᵢ += Δgᵢ
end
return (;
firing_rates,
inputs,
on_spike_arrival!,
Nₑ,
)
end;
using SpikeWorks: Simulation, step!, run!, unpack, newsim,
get_new_spikes!, next_spike, index_of_next
new(; kw...) = begin
ip = input(; kw...)
s = newsim(coba_izh_neuron, ip.inputs, ip.on_spike_arrival!, Δt)
(sim=s, input=ip)
end;
s0 = new().sim
Simulation{Nto1System{NeuronModel{NamedTuple{(:v, :u, :gₑ, :gᵢ), NTuple{4, Float64}}, typeof(f!), typeof(has_spiked), typeof(on_self_spike!)}, var"#on_spike_arrival!#7"{Float64, Float64, var"#neuron_type#6"{Int64}}}, CVec{(:v, :u, :gₑ, :gᵢ)}}
Summary: not started
Properties:
system: Nto1System, x₀: (v = -0.05, u = 0, gₑ = 0, gᵢ = 0), input feed: 0/299503 spikes processed
Δt: 0.0001
duration: 600
stepcounter: 0/6000000
state: t = 0 seconds, neuron = vars: (v: -0.05, u: 0, gₑ: 0, gᵢ: 0), Dₜvars: (v: 0, u: 0, gₑ: 0, gᵢ: 0)
rec:
v: [0, 0, 0, 0, 0, 0, 0, 0, 0, 0 … 0, 0, 0, 0, 0, 0, 0, 0, 0, 0], spiketimes: Float64[]
s0.system
Nto1System{NeuronModel{NamedTuple{(:v, :u, :gₑ, :gᵢ), NTuple{4, Float64}}, typeof(f!), typeof(has_spiked), typeof(on_self_spike!)}, var"#on_spike_arrival!#7"{Float64, Float64, var"#neuron_type#6"{Int64}}}
Summary: Nto1System, x₀: (v = -0.05, u = 0, gₑ = 0, gᵢ = 0), input feed: 0/299503 spikes processed
Properties:
neuronmodel: vars_t₀: (v: -0.05, u: 0, gₑ: 0, gᵢ: 0), f!: f!, has_spiked: has_spiked, on_self_spike!: on_self_spike!
input: 0/299503 spikes processed
on_spike_arrival!: Δgᵢ: 2.4E-09, Δgₑ: 6E-10, neuron_type:
(Nₑ: 80)
(Look at that parametrization of on_spike_arrival! closure :OO)
Sim¶
@time s = run!(new().sim)
2.046088 seconds (12.97 M allocations: 1.022 GiB, 19.90% gc time, 16.45% compilation time)
Simulation{Nto1System{NeuronModel{NamedTuple{(:v, :u, :gₑ, :gᵢ), NTuple{4, Float64}}, typeof(f!), typeof(has_spiked), typeof(on_self_spike!)}, var"#on_spike_arrival!#7"{Float64, Float64, var"#neuron_type#6"{Int64}}}, CVec{(:v, :u, :gₑ, :gᵢ)}}
Summary: completed. 2 spikes/s
Properties:
system: Nto1System, x₀: (v = -0.05, u = 0, gₑ = 0, gᵢ = 0), input feed: all 267219 spikes processed
Δt: 0.0001
duration: 600
stepcounter: 6000000 (complete)
state: t = 600 seconds, neuron = vars: (v: -0.0587, u: 3.52E-12, gₑ: 7.32E-10, gᵢ: 9.52E-10), Dₜvars: (v: 0.0289, u: -1.82E-10, gₑ: -1.06E-07, gᵢ: -1.38E-07)
rec: v: [-0.0501, -0.0501, -0.0502, -0.0503, -0.0503, -0.0504, -0.0505, -0.0506, -0.0506, -0.0507 … -0.0588, -0.0588, -0.0588, -0.0588, -0.0587, -0.0587, -0.0587, -0.0587, -0.0587, -0.0587], spiketimes: [0.429, 0.91, 1.35, 2.62, 2.8, 3.02, 3.31, 3.53, 3.72, 4.2 … 596, 597, 597, 597, 598, 598, 598, 598, 599, 600]
(So 3.3 seconds for 10 minute simulation with N=100 inputs)
Plot¶
v_rec = s.rec.v;
Nt = s.stepcounter.N;
@time using PyPlot
0.001532 seconds (348 allocations: 21.750 KiB)
t = linspace(0, sim_duration, Nt)
plotsig(t, v_rec / mV; tlim=[0,10seconds]);
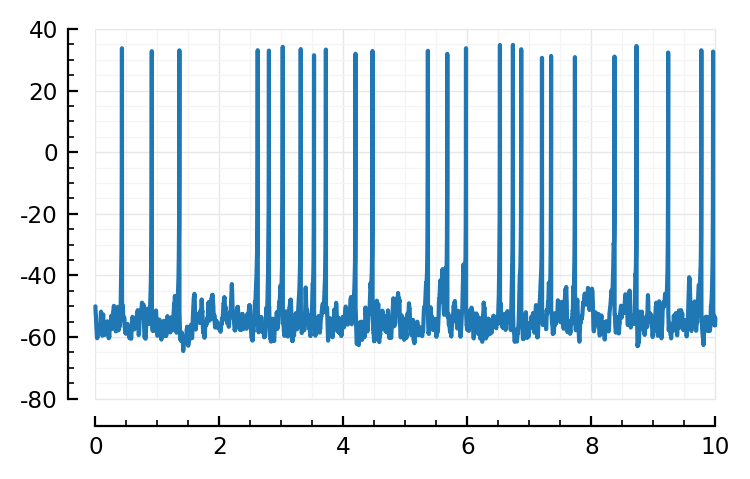
Multi sim¶
(These Ns are same as in e.g. https://tfiers.github.io/phd/nb/2022-10-11__Nto1_output_rate__Edit_of_2022-05-02.html)
using SpikeWorks: spikerate
sim_duration/minutes
10
using Printf
print_Δt(t0) = @printf("%.2G seconds\n", time()-t0)
macro time′(ex) :( t0=time(); $(esc(ex)); print_Δt(t0) ) end;
Ns_and_scalings = [
(5, 2.4), # => N_inh = 1
(20, 1.3),
# orig: 21.
# But: "pₑ = 0.8 does not divide N = 21 into integer parts"
# So voila
(100, 0.8),
(400, 0.6),
(1600, 0.5),
(6500, 0.5),
];
Ns = first.(Ns_and_scalings)
simruns = []
for (N, f) in Ns_and_scalings
scaling = f*N
(sim, inp) = new(; N, scaling)
@show N
@time′ run!(sim)
@show spikerate(sim)
push!(simruns, (; sim, input=inp))
println()
end
N = 5
2.1 seconds
spikerate(sim) = 1.94
N = 20
2.4 seconds
spikerate(sim) = 1.48
N = 100
2.2 seconds
spikerate(sim) = 3.94
N = 400
2.2 seconds
spikerate(sim) = 5.2
N = 1600
3.7 seconds
spikerate(sim) = 5.02
N = 6500
5 seconds
spikerate(sim) = 5.58
Disentangle¶
weird old code. who wrote this?! (oh me)
what the hell is that naming. “input.inputs”
inp = simruns[1].input
st1 = inp.inputs[1].train.spiketimes;
spiketimes(input::Nto1Input) = input.train.spiketimes;
s = simruns[1].sim
s.rec.v;
vrec(s::Simulation{<:Nto1System}) = s.rec.v;
Conntest¶
function conntest_all(inputs, sim)
f(input) = conntest(spiketimes(input), sim)
@showprogress map(f, inputs)
end
conntest_all(simrun) = conntest_all(simrun.input.inputs, simrun.sim);
winsize = 1000
calcSTA(sim, spiketimes) =
calc_STA(vrec(sim), spiketimes, sim.Δt, winsize)
function conntest(spiketimes, sim)
sta = calcSTA(sim, spiketimes)
shufs = [
calcSTA(sim, shuffle_ISIs(spiketimes))
for _ in 1:100
]
test_conn(ptp_test, sta, shufs)
end;
# @code_warntype calc_STA(vrec(s), st1, s.Δt, winsize)
# all good
conntest_all(simruns[1])
Progress: 100%|█████████████████████████████████████████| Time: 0:00:00
5-element Vector{NamedTuple{(:predtype, :pval, :pval_type, :Eness), Tuple{Symbol, Float64, String, Float64}}}:
(predtype = :exc, pval = 0.01, pval_type = "<", Eness = 1.94)
(predtype = :exc, pval = 0.01, pval_type = "<", Eness = 2.19)
(predtype = :exc, pval = 0.01, pval_type = "<", Eness = 2.37)
(predtype = :exc, pval = 0.01, pval_type = "<", Eness = 1.96)
(predtype = :inh, pval = 0.01, pval_type = "<", Eness = -1.28)
conntest_all(simruns[3]);
Progress: 100%|█████████████████████████████████████████| Time: 0:00:25
✔
..but, it takes 25 seconds for simrun 3, i.e. for..
length(simruns[3].input.inputs)
100
..inputs.
so extrapolating, the last one would take
25seconds * 6500/100 / minutes
27.1
Almost half an hour.
So this is why we cached and parallel-processed the STA calculation
Cache STA calc¶
cached()
nbname = "2022-10-24__Nto1_with_fixed_lognormal_inputs"
cachekey(N) = "$(nbname)__N=$N";
cachekey(Ns[end])
"2022-10-24__Nto1_with_fixed_lognormal_inputs__N=6500"
function calc_STA_and_shufs(spiketimes, sim)
realSTA = calcSTA(sim, spiketimes)
shufs = [
calcSTA(sim, shuffle_ISIs(spiketimes))
for _ in 1:100
]
(; realSTA, shufs)
end
"calc_all_STAs_and_shufs"
function calc_all_STAz(inputs, sim)
f(input) = calc_STA_and_shufs(spiketimes(input), sim)
@showprogress map(f, inputs)
end
calc_all_STAz(simrun) = calc_all_STAz(unpakk(simrun)...);
unpakk(simrun) = (; simrun.input.inputs, simrun.sim)
out = calc_all_STAz(simruns[1])
print(Base.summary(out))
Progress: 100%|█████████████████████████████████████████| Time: 0:00:01
5-element Vector{NamedTuple{(:realSTA, :shufs), Tuple{Vector{Float64}, Vector{Vector{Float64}}}}}
calc_all_cached(i) = cached(calc_all_STAz, [simruns[i]], key=cachekey(Ns[i]))
out = []
for i in eachindex(simruns)
push!(out, calc_all_cached(i))
end;
Loading cached output from `C:\Users\tfiers\.phdcache\calc_all_STAz\2022-10-24__Nto1_with_fixed_lognormal_inputs__N=5.jld2` … done (3.0 s)
Loading cached output from `C:\Users\tfiers\.phdcache\calc_all_STAz\2022-10-24__Nto1_with_fixed_lognormal_inputs__N=20.jld2` … done (0.1 s)
Loading cached output from `C:\Users\tfiers\.phdcache\calc_all_STAz\2022-10-24__Nto1_with_fixed_lognormal_inputs__N=100.jld2` … done (5.6 s)
Loading cached output from `C:\Users\tfiers\.phdcache\calc_all_STAz\2022-10-24__Nto1_with_fixed_lognormal_inputs__N=400.jld2` … done (1.1 s)
Loading cached output from `C:\Users\tfiers\.phdcache\calc_all_STAz\2022-10-24__Nto1_with_fixed_lognormal_inputs__N=1600.jld2` … done (5.0 s)
Loading cached output from `C:\Users\tfiers\.phdcache\calc_all_STAz\2022-10-24__Nto1_with_fixed_lognormal_inputs__N=6500.jld2` … done (47.6 s)
path = raw"C:\Users\tfiers\.phdcache\calc_all_STAz\2022-10-24__Nto1_with_fixed_lognormal_inputs__N=6500.jld2"
stat(path).size / GB
5.3
(Yeah, shuffle test not great here)
Conntest based on STA cache¶
[test_conn(ptp_test, sta, shufs) for (sta,shufs) in out[1]]
5-element Vector{NamedTuple{(:predtype, :pval, :pval_type, :Eness), Tuple{Symbol, Float64, String, Float64}}}:
(predtype = :exc, pval = 0.01, pval_type = "<", Eness = 1.94)
(predtype = :exc, pval = 0.01, pval_type = "<", Eness = 2.19)
(predtype = :exc, pval = 0.01, pval_type = "<", Eness = 2.37)
(predtype = :exc, pval = 0.01, pval_type = "<", Eness = 1.96)
(predtype = :inh, pval = 0.01, pval_type = "<", Eness = -1.28)
✔, same as above
Two-stage conntest, ptp-then-correlation¶
..wait, that assumes we can even find some true connections with ptp.
So let’s try that.
# We need.. a column with `conntype`, the real type.
i = last(eachindex(simruns))
6
sim, inp = simruns[i];
Nₑ = inp.Nₑ
5200
N = Ns[i]
6500
conntype_vec(i) = begin
sim, inp = simruns[i]
Nₑ = inp.Nₑ
N = Ns[i]
conntype = Vector{Symbol}(undef, N);
conntype[1:Nₑ] .= :exc
conntype[Nₑ+1:end] .= :inh
conntype
end;
conntestresults(i, teststat = ptp_test; α = 0.05) = begin
f((sta, shufs)) = test_conn(teststat, sta, shufs; α)
res = @showprogress map(f, out[i])
df = DataFrame(res)
df[!, :conntype] = conntype_vec(i)
df
end;
conntestresults(1)
Progress: 100%|█████████████████████████████████████████| Time: 0:00:00
| Row | predtype | pval | pval_type | Eness | conntype |
|---|---|---|---|---|---|
| Symbol | Float64 | String | Float64 | Symbol | |
| 1 | exc | 0.01 | < | 1.94 | exc |
| 2 | exc | 0.01 | < | 2.19 | exc |
| 3 | exc | 0.01 | < | 2.37 | exc |
| 4 | exc | 0.01 | < | 1.96 | exc |
| 5 | inh | 0.01 | < | -1.28 | inh |
ctr = conntestresults(6)
Progress: 100%|█████████████████████████████████████████| Time: 0:00:01
| Row | predtype | pval | pval_type | Eness | conntype |
|---|---|---|---|---|---|
| Symbol | Float64 | String | Float64 | Symbol | |
| 1 | unconn | 0.3 | = | -0.0134 | exc |
| 2 | inh | 0.03 | = | -0.145 | exc |
| 3 | unconn | 0.06 | = | 0.0225 | exc |
| 4 | unconn | 0.84 | = | 0.313 | exc |
| 5 | unconn | 0.7 | = | 0.0951 | exc |
| 6 | unconn | 0.38 | = | -0.15 | exc |
| 7 | unconn | 0.59 | = | -0.0694 | exc |
| 8 | unconn | 0.69 | = | 0.24 | exc |
| 9 | unconn | 0.88 | = | 0.13 | exc |
| 10 | exc | 0.01 | = | 0.567 | exc |
| 11 | unconn | 0.39 | = | 0.0595 | exc |
| 12 | unconn | 0.23 | = | 0.0489 | exc |
| 13 | unconn | 0.2 | = | -0.243 | exc |
| ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ |
| 6489 | unconn | 0.55 | = | -0.0105 | inh |
| 6490 | unconn | 0.64 | = | -0.0366 | inh |
| 6491 | unconn | 0.94 | = | 0.101 | inh |
| 6492 | unconn | 0.57 | = | -0.392 | inh |
| 6493 | unconn | 0.43 | = | 0.198 | inh |
| 6494 | unconn | 0.95 | = | -0.178 | inh |
| 6495 | unconn | 0.18 | = | -0.289 | inh |
| 6496 | unconn | 0.29 | = | -0.0286 | inh |
| 6497 | inh | 0.04 | = | -0.00851 | inh |
| 6498 | unconn | 0.5 | = | 0.206 | inh |
| 6499 | inh | 0.03 | = | -0.0482 | inh |
| 6500 | unconn | 0.48 | = | -0.271 | inh |
Eval¶
pm = perfmeasures(ctr)
perftable(ctr)
| Tested connections: 6500 | ||||||
|---|---|---|---|---|---|---|
| ┌─────── | Real type | ───────┐ | Precision | |||
unconn | exc | inh | ||||
| ┌ | unconn | 0 | 4866 | 1146 | 0% | |
| Predicted type | exc | 0 | 159 | 97 | 62% | |
| └ | inh | 0 | 175 | 57 | 25% | |
| Sensitivity | NaN% | 3% | 4% |
So 3 to 4% of connections detected.
α = FPR = 5%.
So, alas
Analyse¶
Did the high firing inputs fare better?
sim,inp = simruns[6]
inp.firing_rates;
For starters, are the input firing rates the actual firing rates:
spikerate_(spiketimes) = length(spiketimes) / sim_duration;
using Sciplotlib: plot
stipulated_firing_rates = inp.firing_rates
real_firing_rates = spikerate_.(spiketimes.(inp.inputs))
plot(stipulated_firing_rates, real_firing_rates);
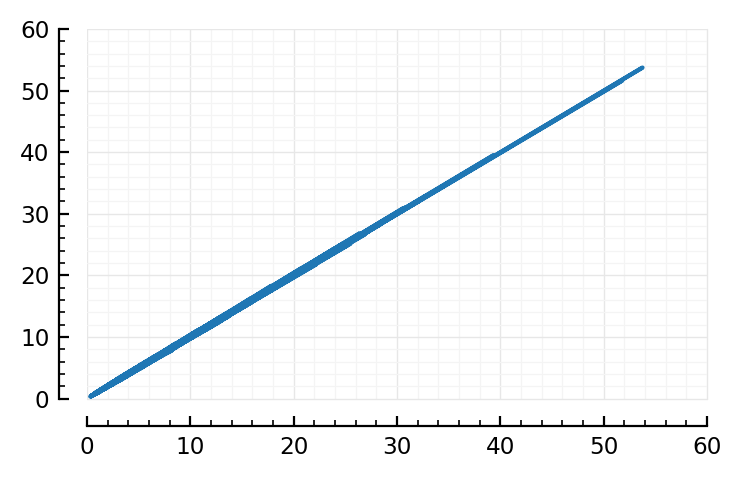
Ok, check
fr, nid = findmax(real_firing_rates)
(53.7, 6192)
(It’s an inh one)
plotSTA(nid) = plot(calcSTA(sim, spiketimes(inp.inputs[nid])) / mV);
plotSTA(nid);
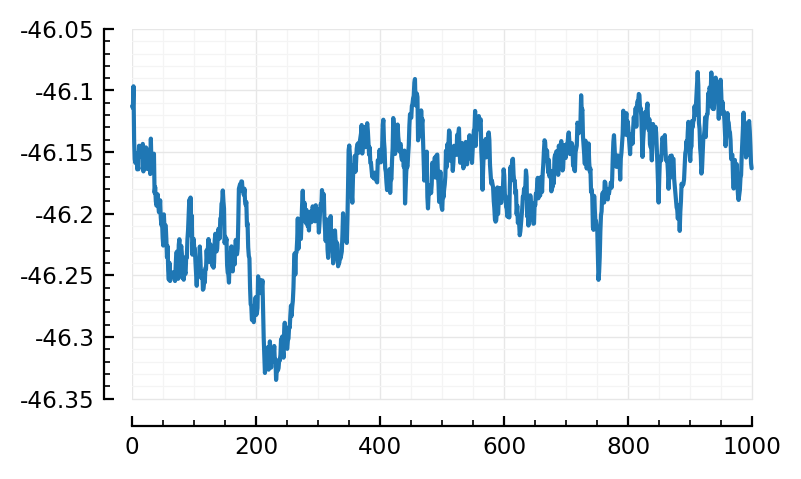
fr, nid_exc = findmax(real_firing_rates[1:inp.Nₑ])
(51.6, 1431)
plotSTA(nid_exc);
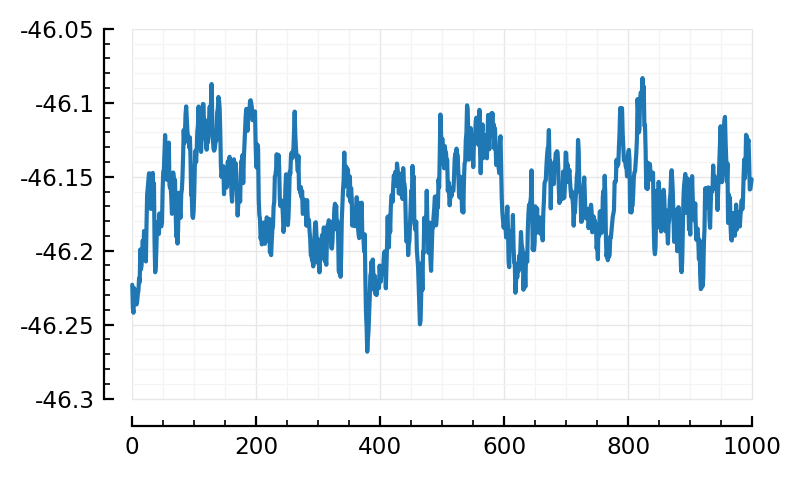
Alas alas.
What about the second to last one, N = 1600¶
i = length(Ns) - 1
5
Ns[i]
1600
sims = first.(simruns);
inps = last.(simruns);
firing_rates(i) = spikerate_.(spiketimes.(inps[i].inputs))
fr,ni = findmax(firing_rates(5))
(36.5, 434)
plotSTA(i,ni) = plot(calcSTA(sims[i], spiketimes(inps[i].inputs[ni])) / mV);
plotSTA(i,ni);
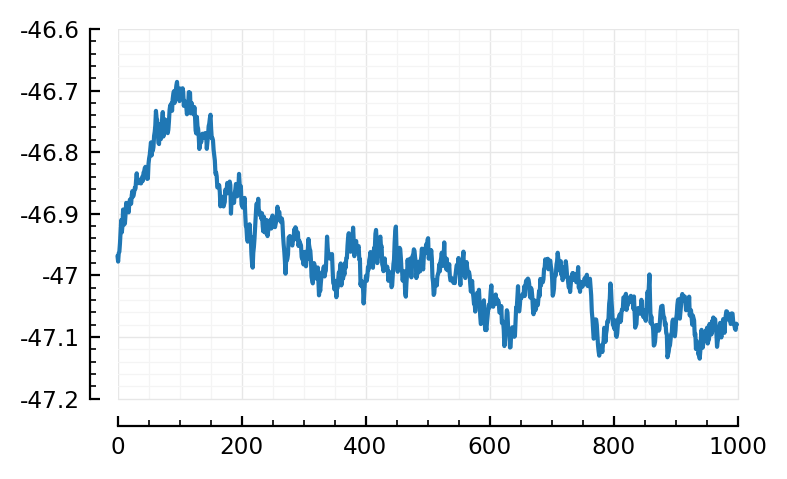
Ah, that’s better!
ctr = conntestresults(5)
perftable(ctr)
Progress: 100%|█████████████████████████████████████████| Time: 0:00:00
| Tested connections: 1600 | ||||||
|---|---|---|---|---|---|---|
| ┌─────── | Real type | ───────┐ | Precision | |||
unconn | exc | inh | ||||
| ┌ | unconn | 0 | 1063 | 131 | 0% | |
| Predicted type | exc | 0 | 112 | 74 | 60% | |
| └ | inh | 0 | 105 | 115 | 52% | |
| Sensitivity | NaN% | 9% | 36% |
So here it’s worth doing a two-pass test:
Two-pass test (strict ptp, then correlation)¶
ctr_strict = conntestresults(5, α=1/100)
perftable(ctr_strict)
Progress: 100%|█████████████████████████████████████████| Time: 0:00:00
| Tested connections: 1600 | ||||||
|---|---|---|---|---|---|---|
| ┌─────── | Real type | ───────┐ | Precision | |||
unconn | exc | inh | ||||
| ┌ | unconn | 0 | 1171 | 166 | 0% | |
| Predicted type | exc | 0 | 56 | 64 | 47% | |
| └ | inh | 0 | 53 | 90 | 63% | |
| Sensitivity | NaN% | 4% | 28% |
ids = findall(ctr_strict.predtype .== :exc)
length(ids)
120
Hm although: of the 120 connections predicted ‘exc’, more than half are actually inh
Let’s see what an average STA gives anyway
sim,inp = simruns[5]
STAs_predicted_exc = [calcSTA(sim, spiketimes(inp.inputs[i])) for i in ids];
template = mean(STAs_predicted_exc)
plot(template/mV);
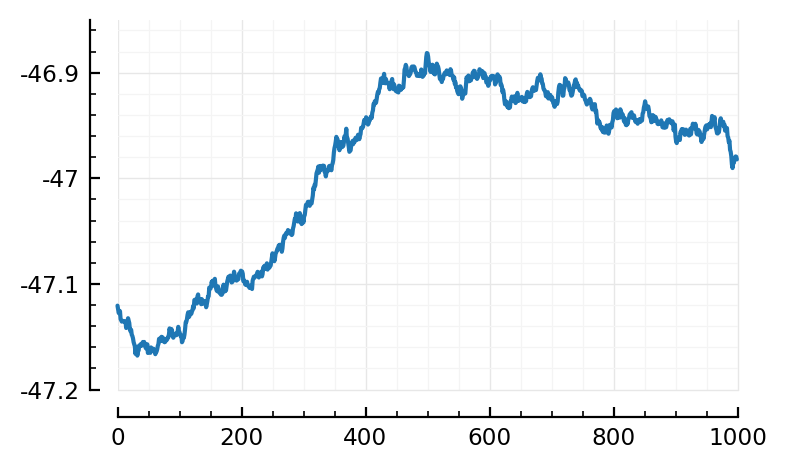
Hm, interesting. Not the previously known STA shape.
[not furhter explored why]
Now to correlation-conntest with this
ctr2 = conntestresults(5, corr_test $ (; template), α = 0.05)
perftable(ctr2)
Progress: 100%|█████████████████████████████████████████| Time: 0:00:00
| Tested connections: 1600 | ||||||
|---|---|---|---|---|---|---|
| ┌─────── | Real type | ───────┐ | Precision | |||
unconn | exc | inh | ||||
| ┌ | unconn | 0 | 922 | 84 | 0% | |
| Predicted type | exc | 0 | 12 | 236 | 5% | |
| └ | inh | 0 | 346 | 0 | 0% | |
| Sensitivity | NaN% | 1% | 0% |
Lol it’s worse.
Ok, that’s cause there’s more inh in template.
(That’s why STA lookd different)
So let’s use inh as template
ids_inh = findall(ctr_strict.predtype .== :inh)
length(ids_inh)
143
STAs_predicted_inh = [calcSTA(sim, spiketimes(inp.inputs[i])) for i in ids_inh];
plot(mean(STAs_predicted_inh)/mV)
template_inh = - mean(STAs_predicted_inh); # Note the minus
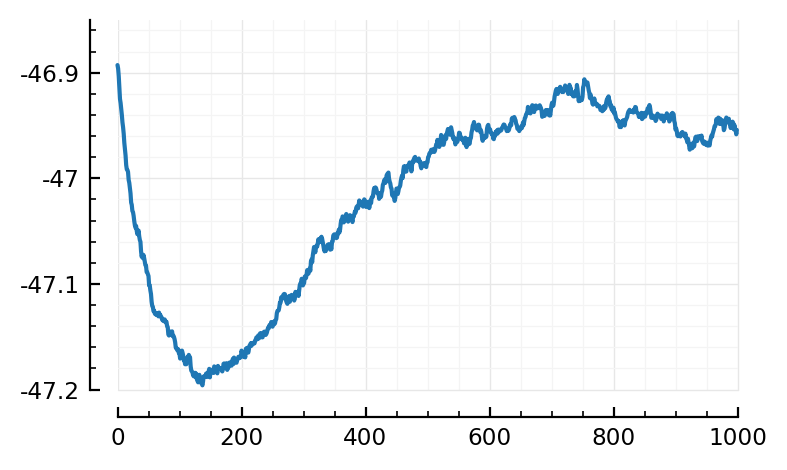
That’s more like it
ctr3 = conntestresults(5, corr_test $ (; template=template_inh), α = 0.05)
perftable(ctr3)
Progress: 100%|█████████████████████████████████████████| Time: 0:00:00
| Tested connections: 1600 | ||||||
|---|---|---|---|---|---|---|
| ┌─────── | Real type | ───────┐ | Precision | |||
unconn | exc | inh | ||||
| ┌ | unconn | 0 | 842 | 58 | 0% | |
| Predicted type | exc | 0 | 429 | 0 | 100% | |
| └ | inh | 0 | 9 | 262 | 97% | |
| Sensitivity | NaN% | 34% | 82% |
Ok, not bad :)
Comparing with prev results here: https://tfiers.github.io/phd/nb/2022-04-28__interpolate_N_from_30_to_6000.html#plot-results
At 1600 inputs, there TPRₑ was 5% (here 34%)
and TPRᵢ was 21% (here 82%)
ofc that was with just ptp test. this is with the two phase test.
Just the ptp here:
TPRₑ 9%
TPRᵢ 36%
so it is a bit better, with the lognormal input firing
Try two-pass test on N=6500 anyway¶
But, as above, with the inh as template.
ctr6_strict = conntestresults(6, α=1/100)
perftable(ctr6_strict)
Progress: 100%|█████████████████████████████████████████| Time: 0:00:24
| Tested connections: 6500 | ||||||
|---|---|---|---|---|---|---|
| ┌─────── | Real type | ───────┐ | Precision | |||
unconn | exc | inh | ||||
| ┌ | unconn | 0 | 5064 | 1232 | 0% | |
| Predicted type | exc | 0 | 70 | 45 | 61% | |
| └ | inh | 0 | 66 | 23 | 26% | |
| Sensitivity | NaN% | 1% | 2% |
ids6_inh = findall(ctr6_strict.predtype .== :inh)
length(ids6_inh)
89
sim,inp = simruns[6]
STAs6_predicted_inh = [calcSTA(sim, spiketimes(inp.inputs[i]))
for i in ids6_inh];
avg = mean(STAs6_predicted_inh)
plot(avg/mV)
template_inh6 = - avg;
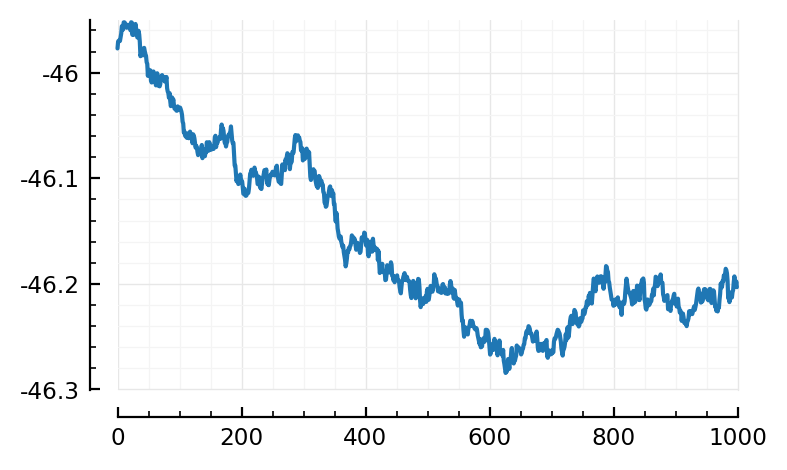
Hm. Less convincing.
(Let’s try anyway)
ctr6_2 = conntestresults(
6, corr_test $ (; template=template_inh6), α = 0.05
)
perftable(ctr6_2)
Progress: 100%|█████████████████████████████████████████| Time: 0:00:01
| Tested connections: 6500 | ||||||
|---|---|---|---|---|---|---|
| ┌─────── | Real type | ───────┐ | Precision | |||
unconn | exc | inh | ||||
| ┌ | unconn | 0 | 4521 | 1021 | 0% | |
| Predicted type | exc | 0 | 180 | 266 | 40% | |
| └ | inh | 0 | 499 | 13 | 3% | |
| Sensitivity | NaN% | 3% | 1% |
Just ptp above (H2 ‘Eval’) had
Precision
62%
25%
Sensitivity 3% 4%
So, this is worse.