2021-01-13 • Multiple ROC’s
Contents
2021-01-13 • Multiple ROC’s¶
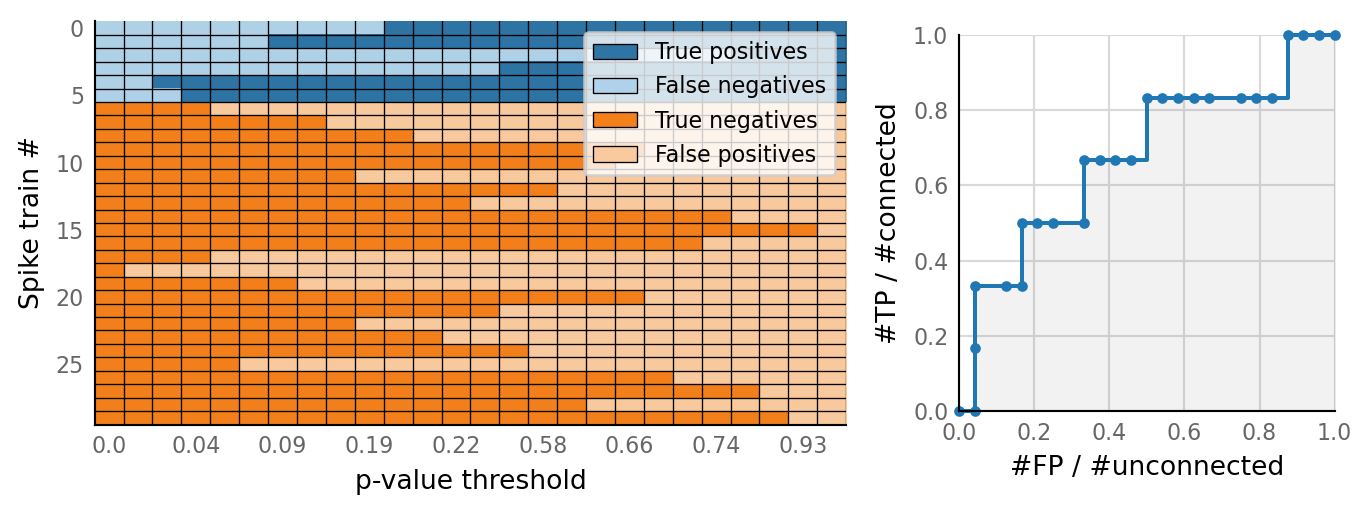
Here we repeat the simulation multiple times (each time varying one parameter), and for each repetition calculate an ROC curve (and associated area under that curve): how well could we classify all (spike_train, VI_signal)-pairs as connected or unconnected, for a range of p-value thresholds?
Setup¶
from voltage_to_wiring_sim.notebook_init import *
v.print_reproducibility_info()
This cell was last run by tfiers on yoga on Tue 26 Jan 2021, at 00:13 (UTC+0100).
Last git commit (Tue 26 Jan 2021, 00:07).
No uncommitted changes.
from voltage_to_wiring_sim.N_to_1_experiment import N_to_1_SimParams, simulate, plot_sim_slice
from voltage_to_wiring_sim.sim.neuron_params import cortical_RS
Base parameters¶
base_params = N_to_1_SimParams(
sim_duration = 1 * minute,
timestep = 0.1 * ms,
num_spike_trains = 30,
p_connected = 0.5,
spike_rate = 20 * Hz,
Δg_syn = 0.8 * nS,
τ_syn = 7 * ms,
neuron_params = cortical_RS,
imaging_spike_SNR = 2,
rng_seed=1800,
);
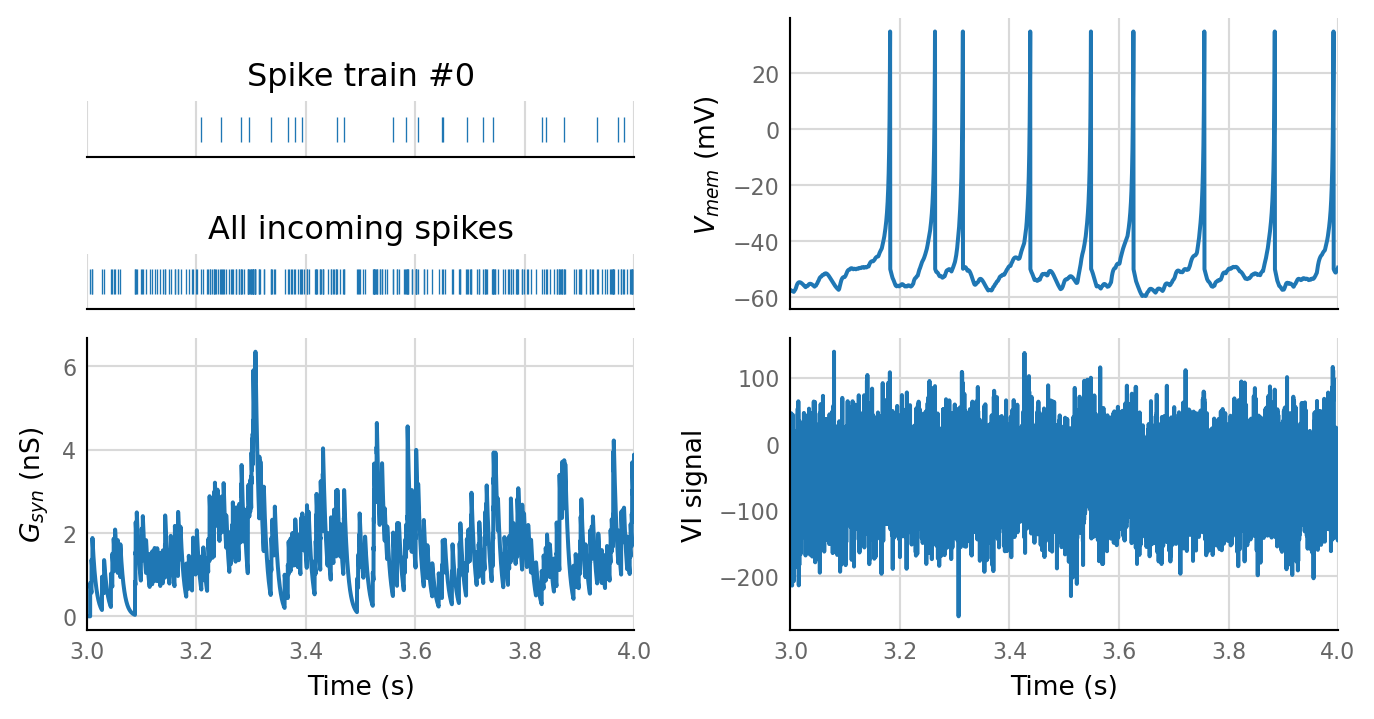
Note the medium short recording and low imaging SNR.
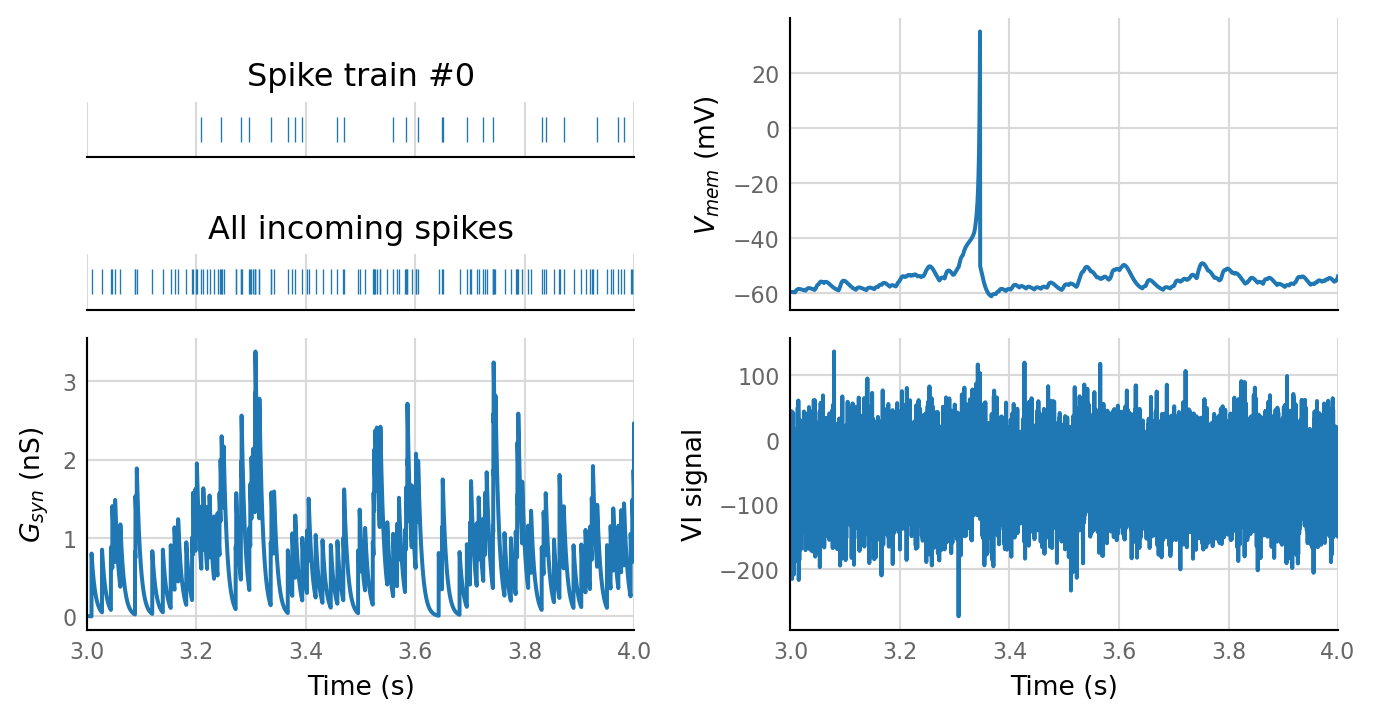
sim_data = simulate(base_params);
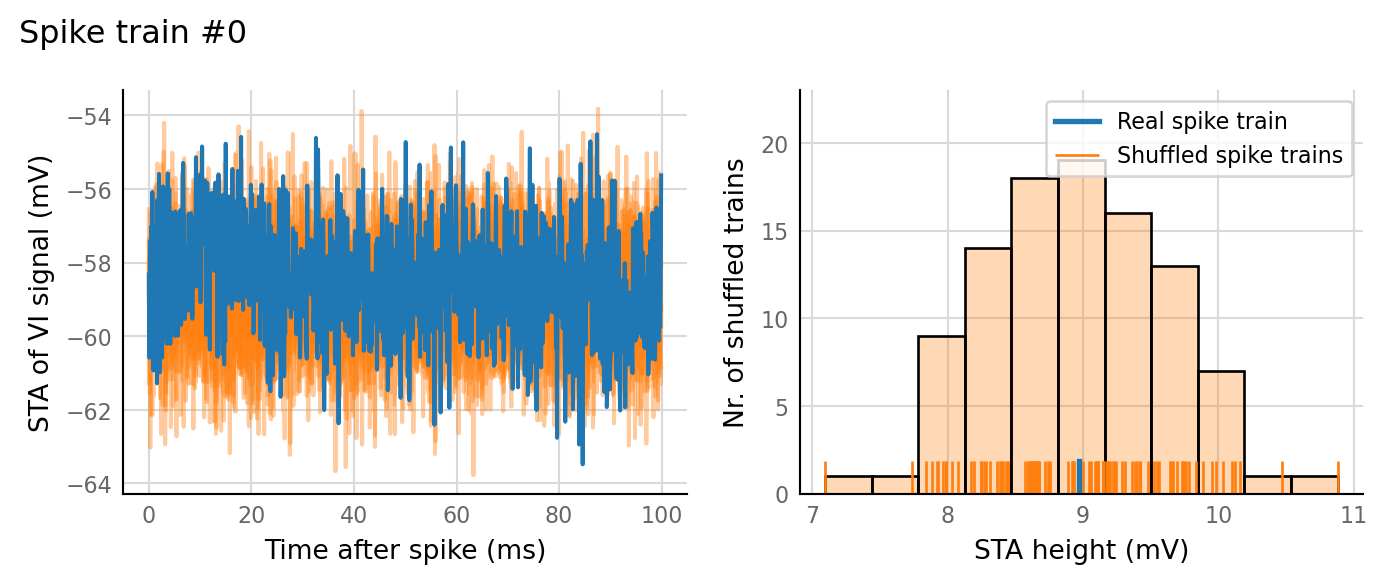
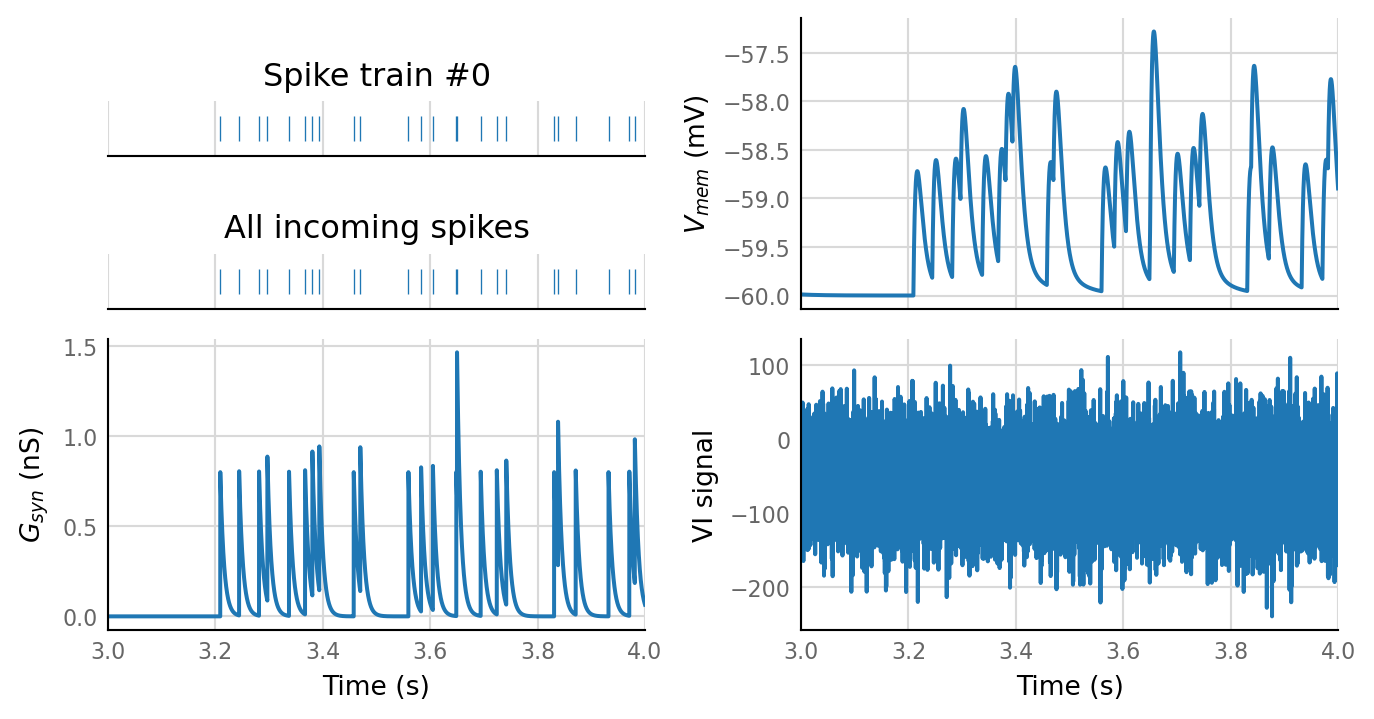
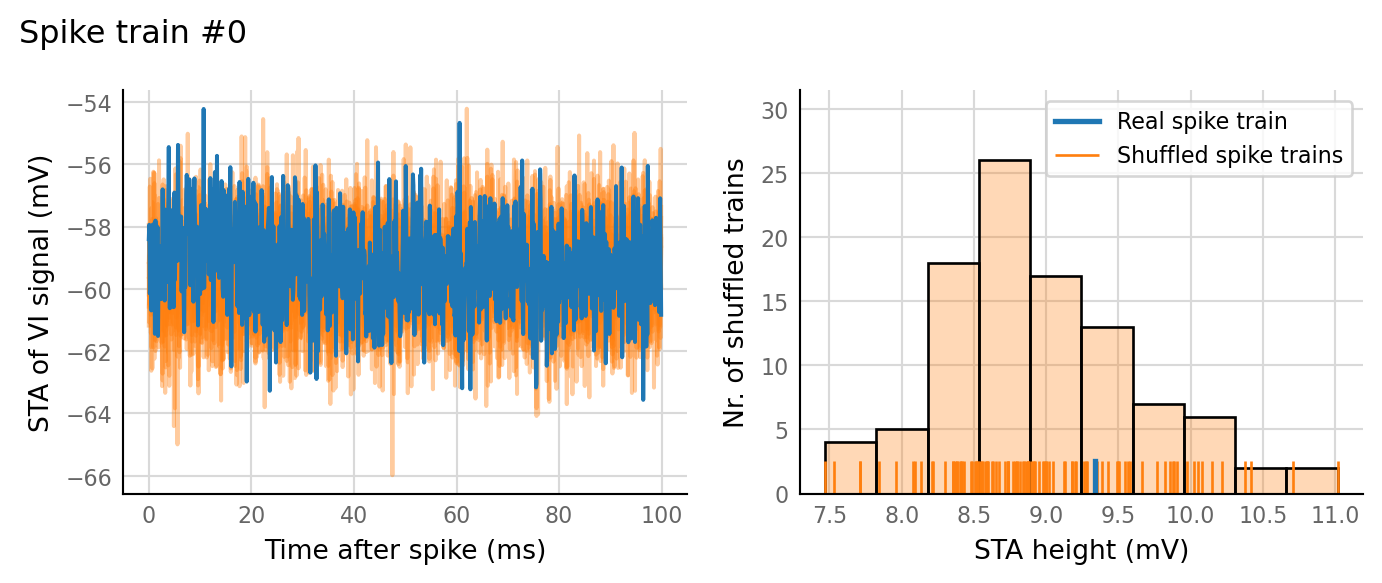
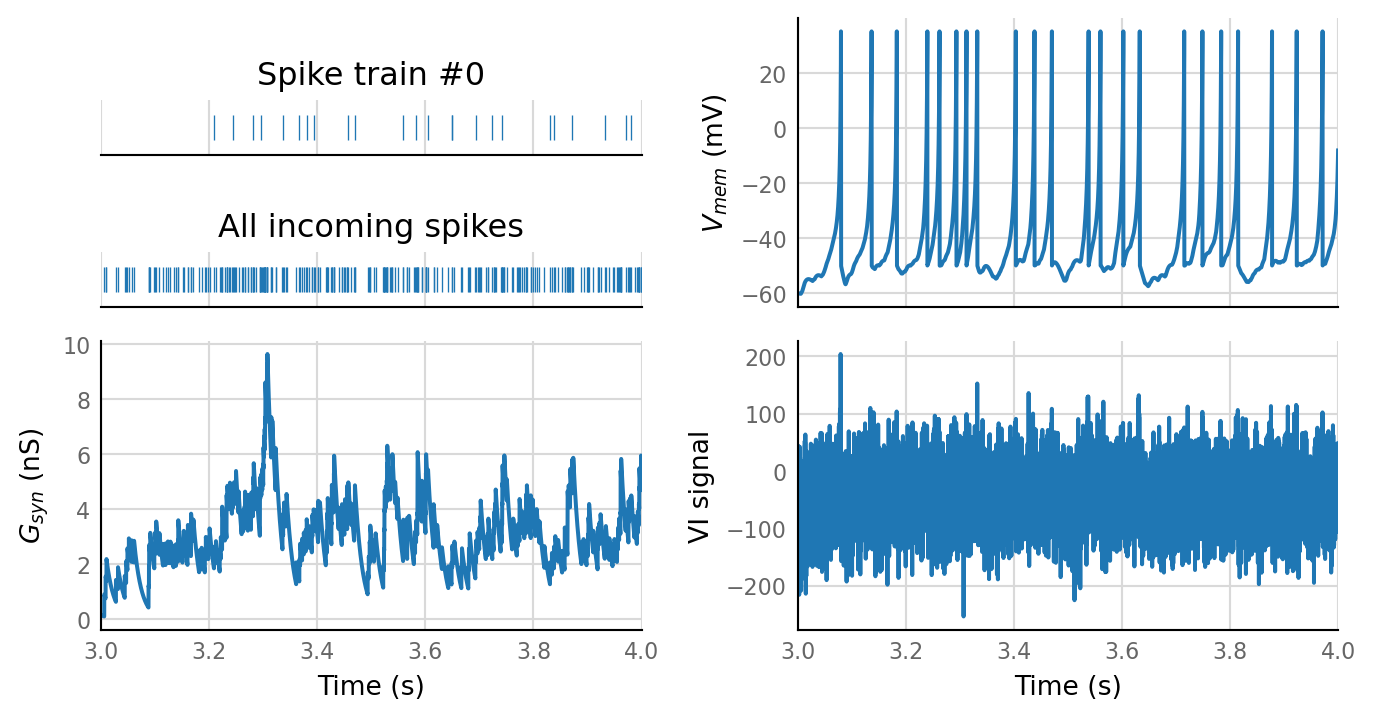
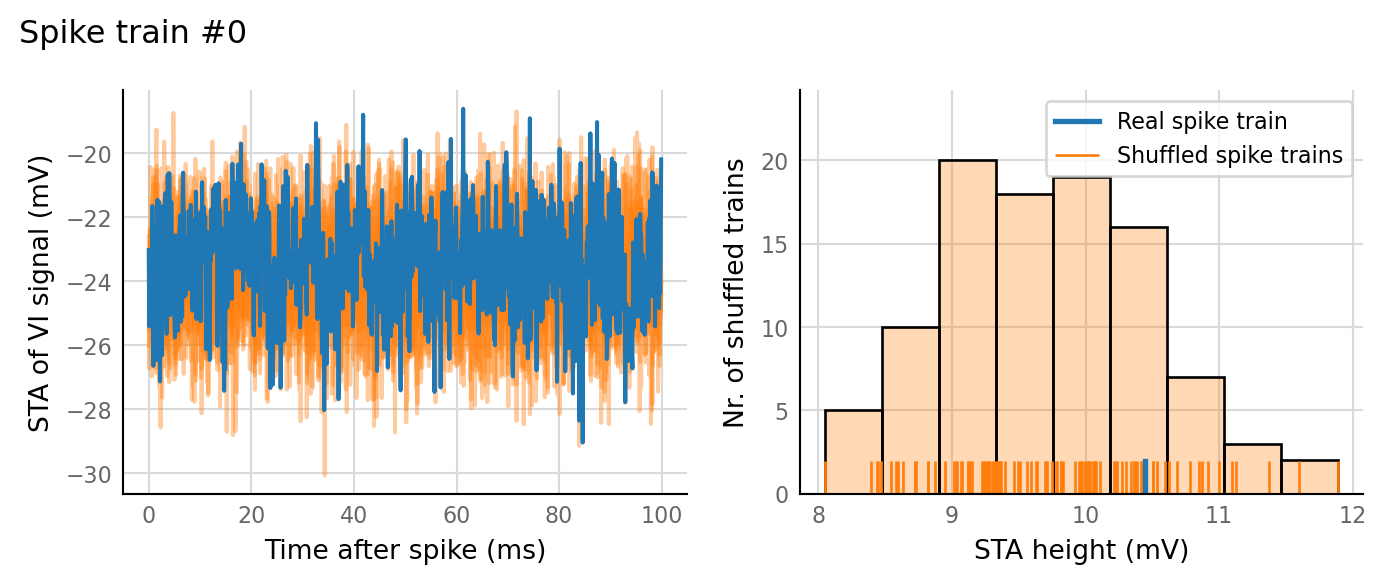
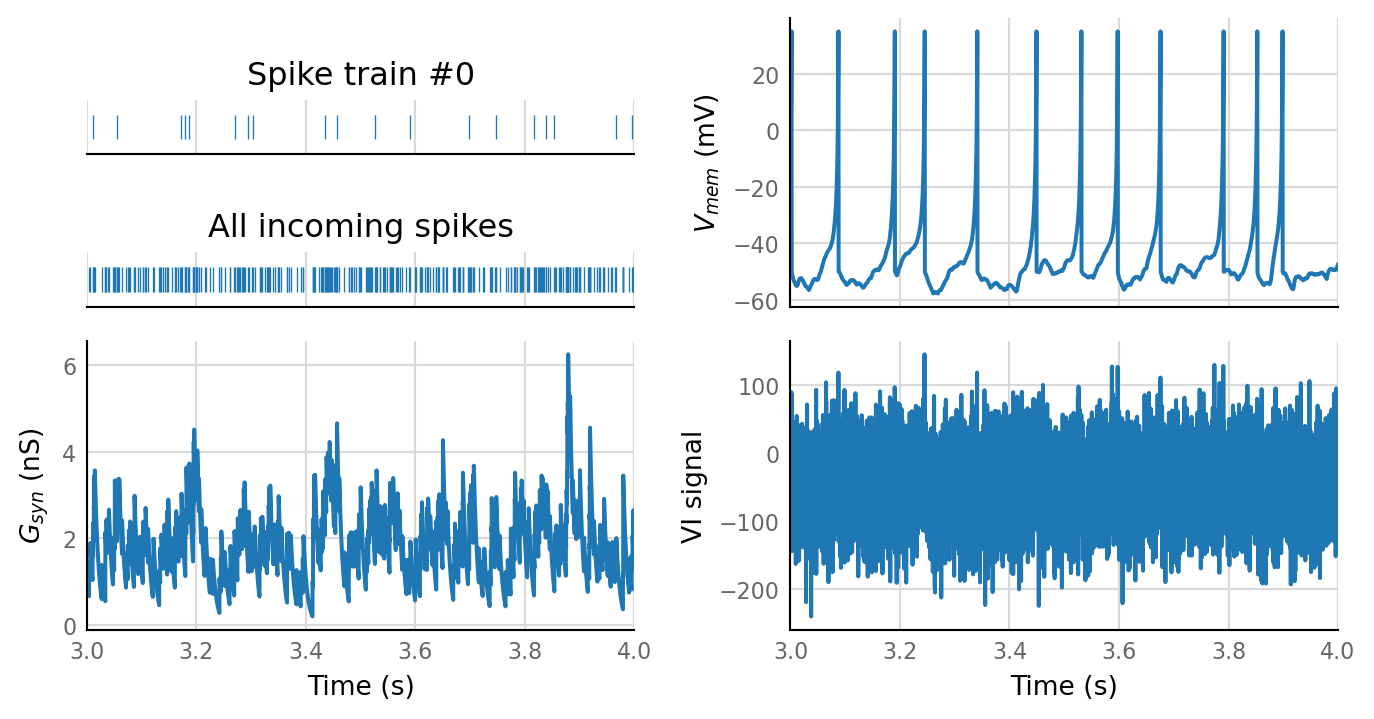
plot_sim_slice(sim_data, t_start=3 * second, duration=1 * second);
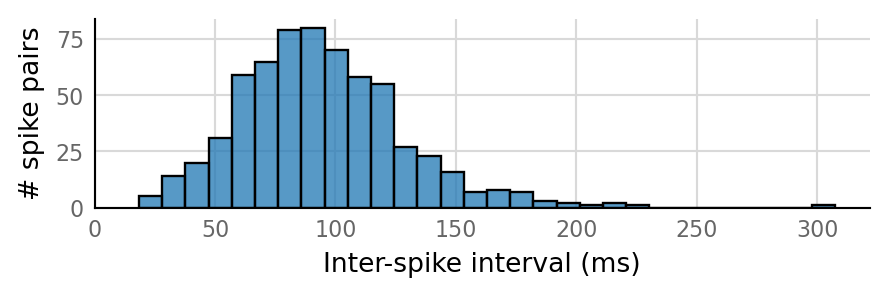
v.sim.izhikevich_neuron.show_output_spike_stats(sim_data.izh_output);
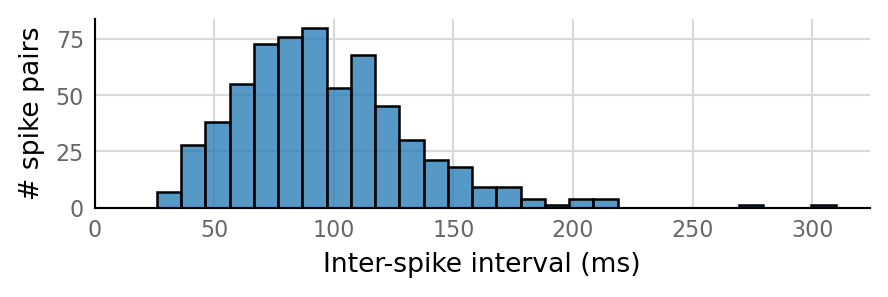
Output spike rate (1 / median ISI): 11 Hz
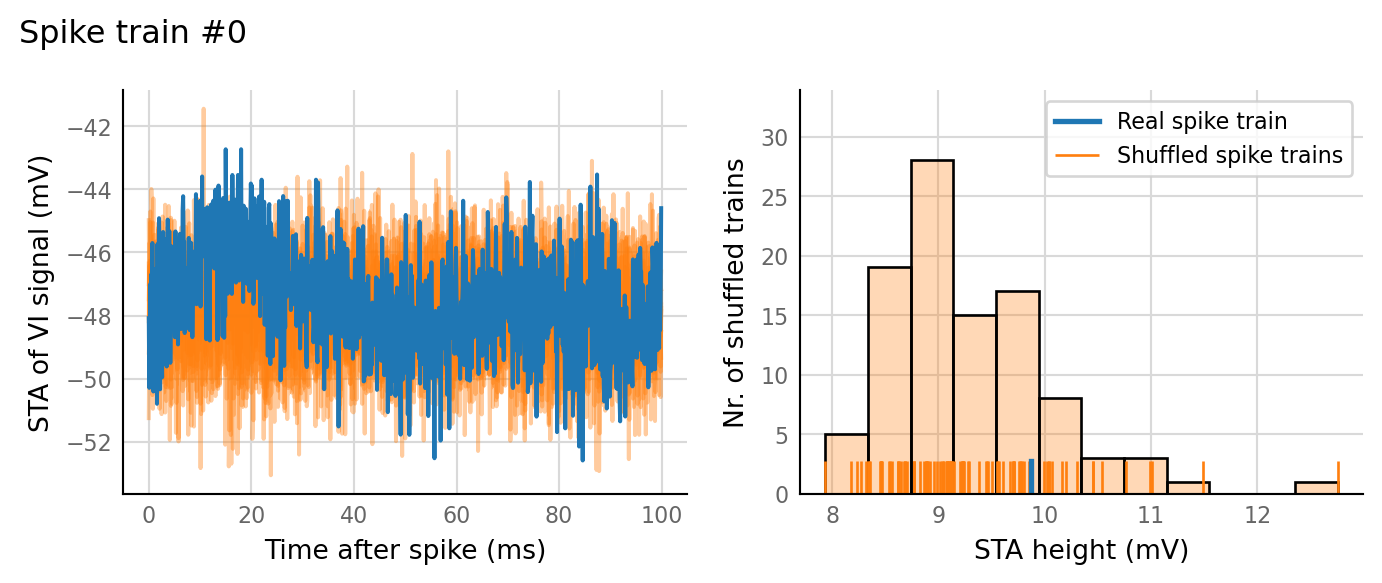
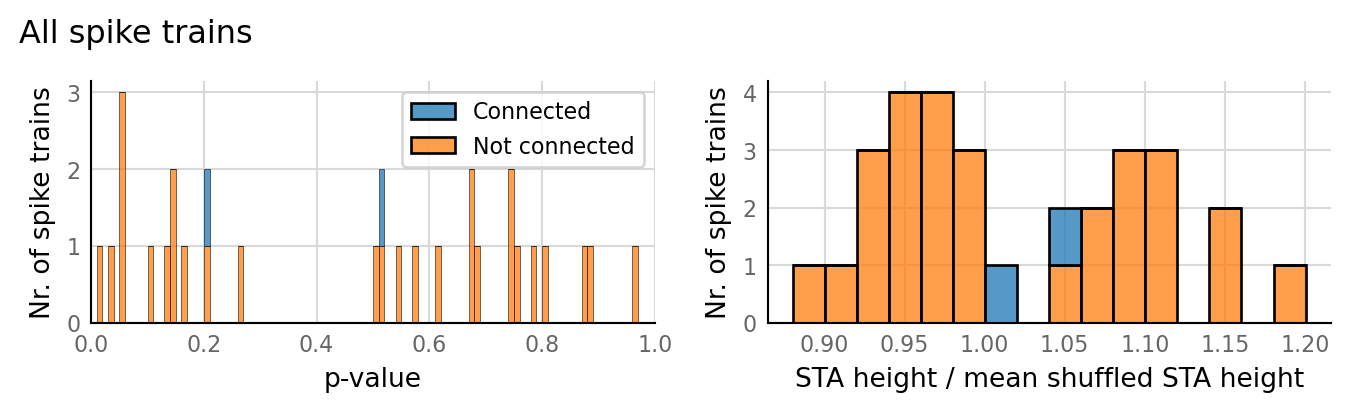
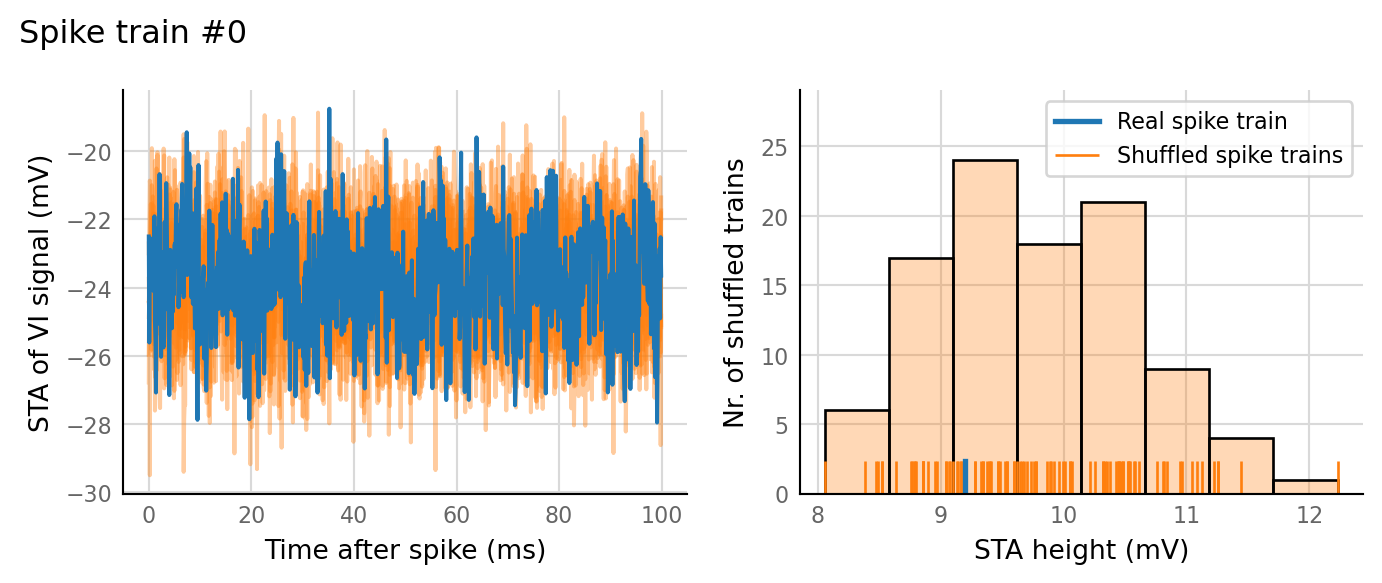
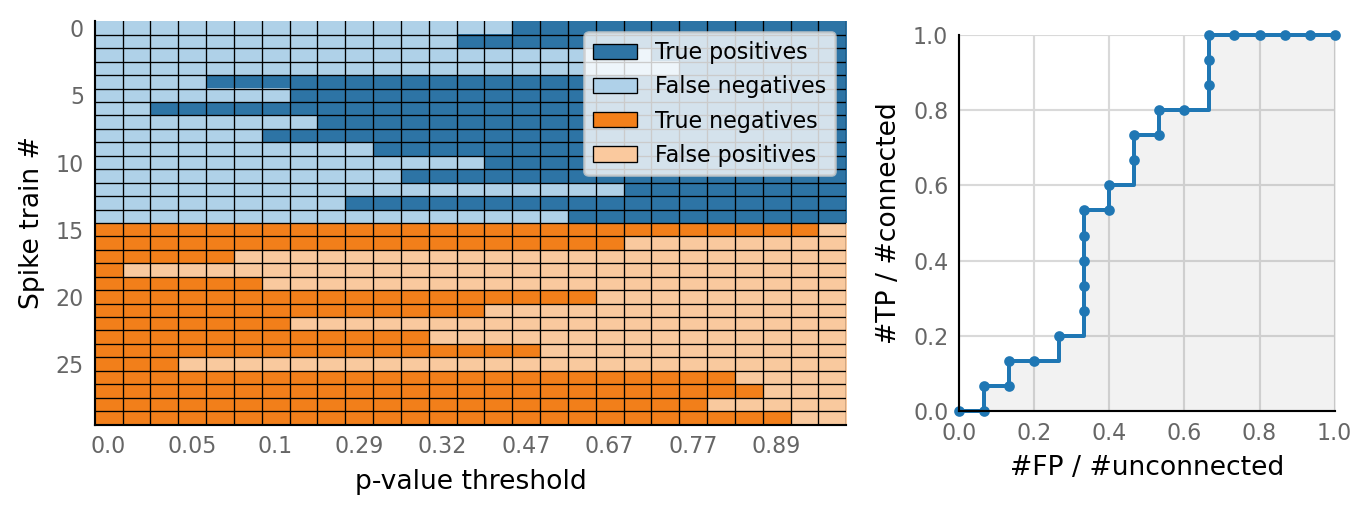
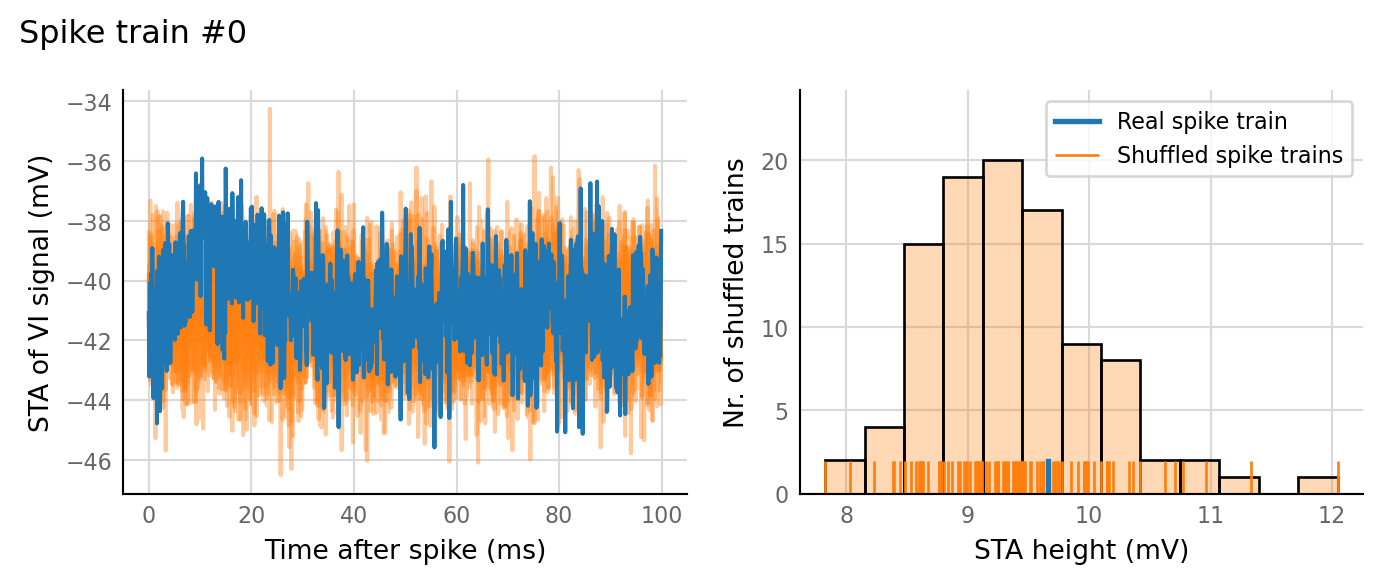
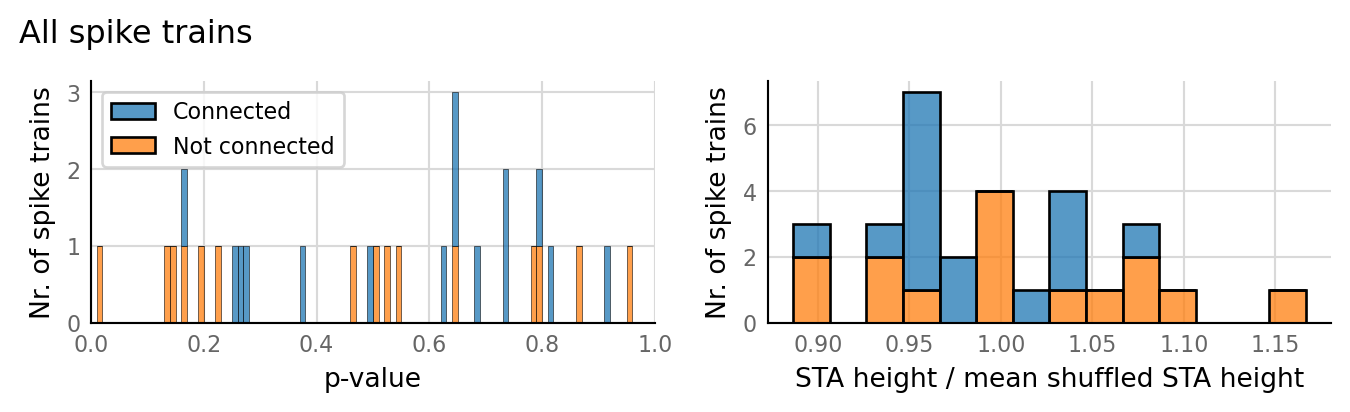
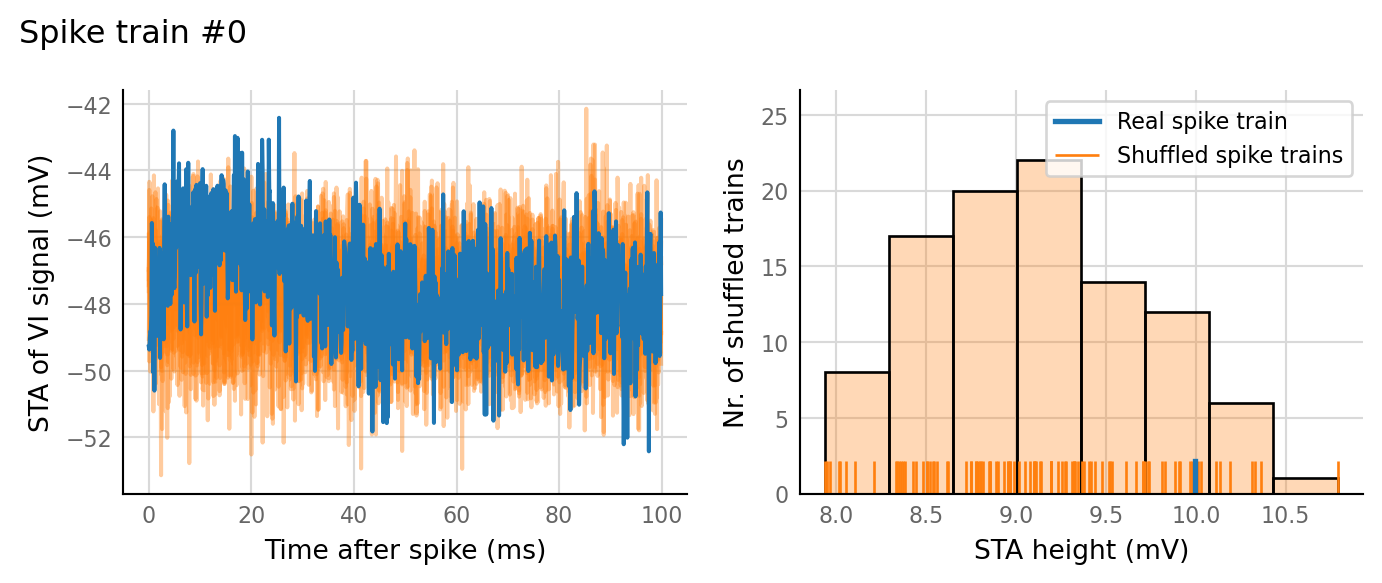
test_data, test_summaries = v.N_to_1_experiment.test_connections(sim_data);
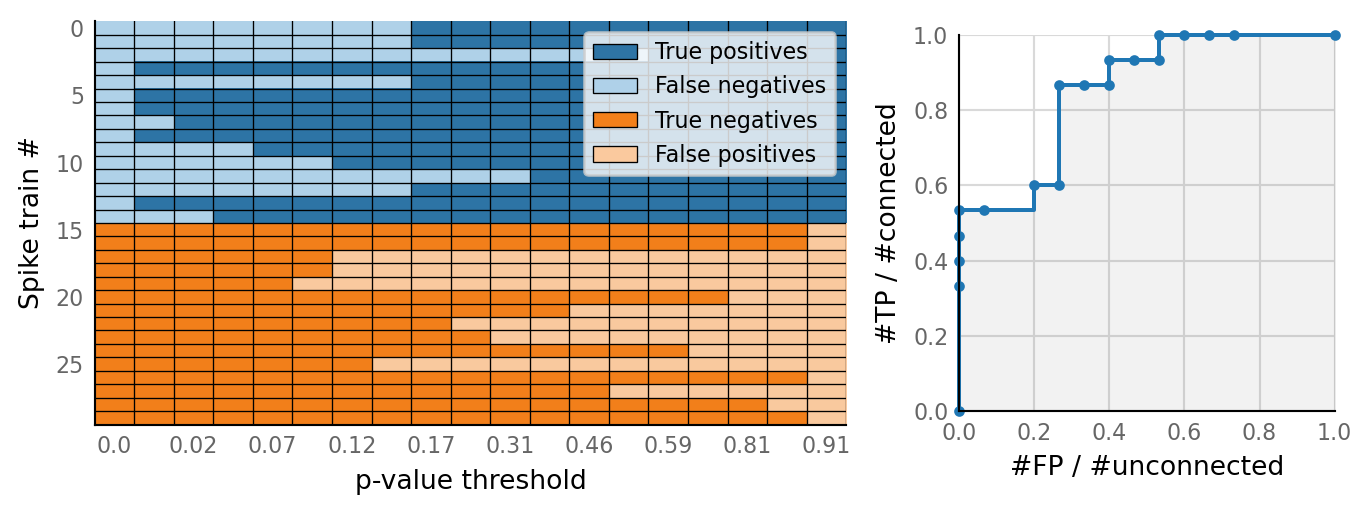
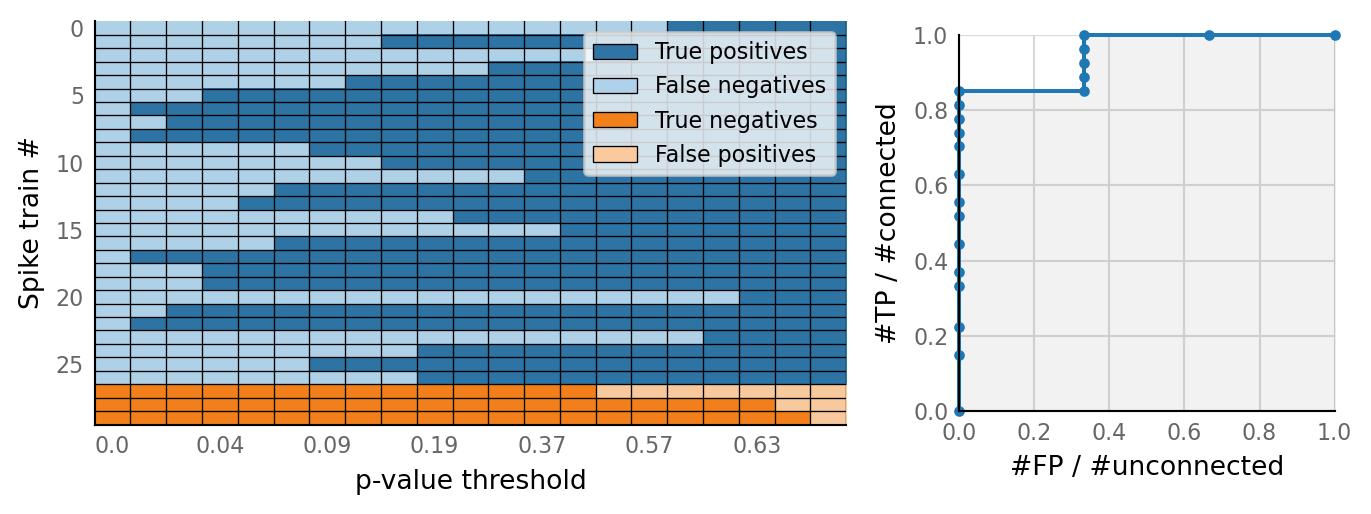
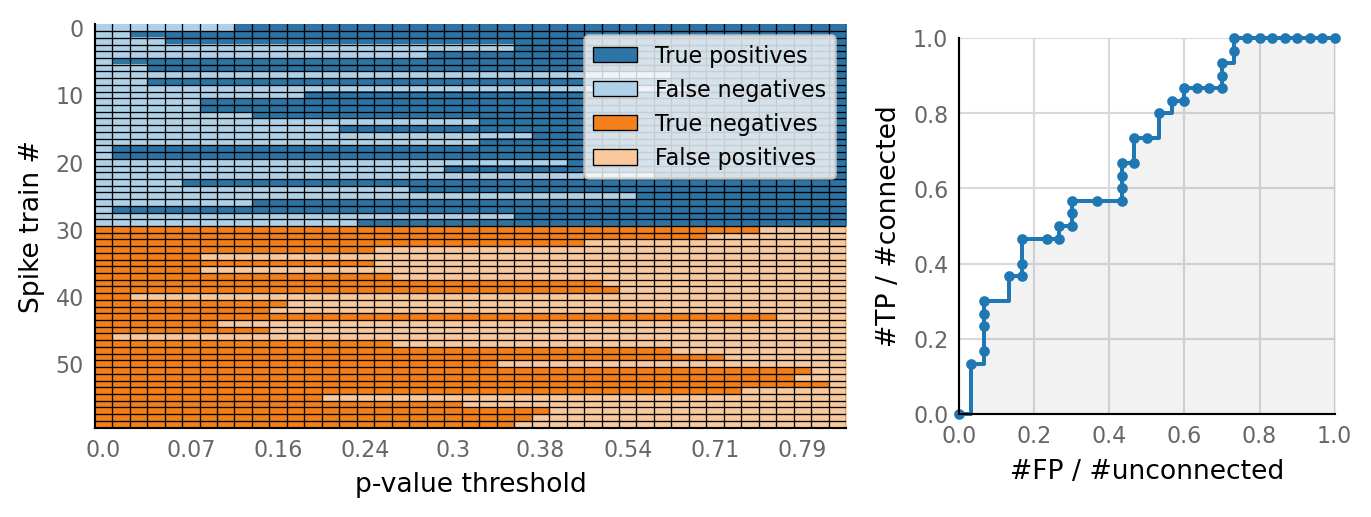
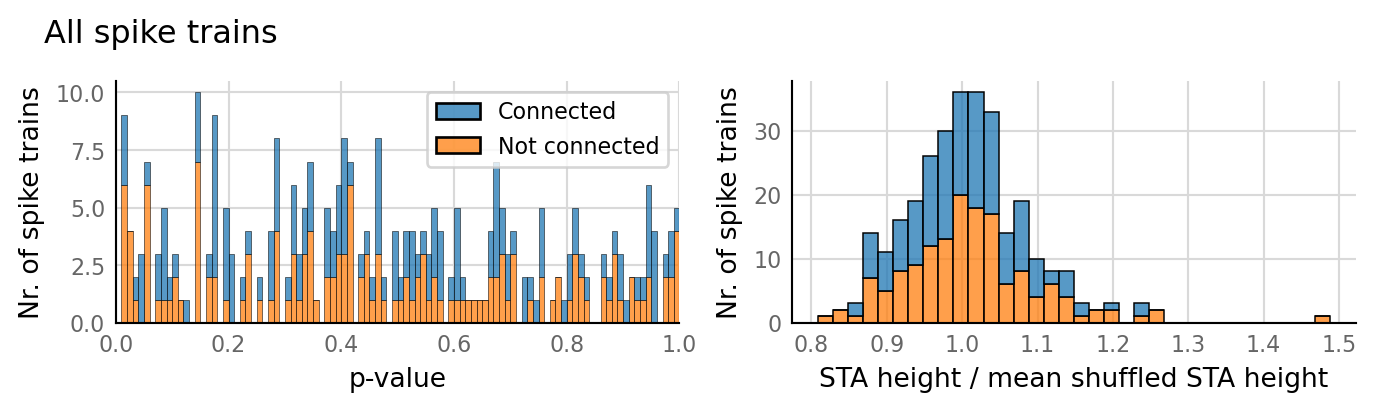
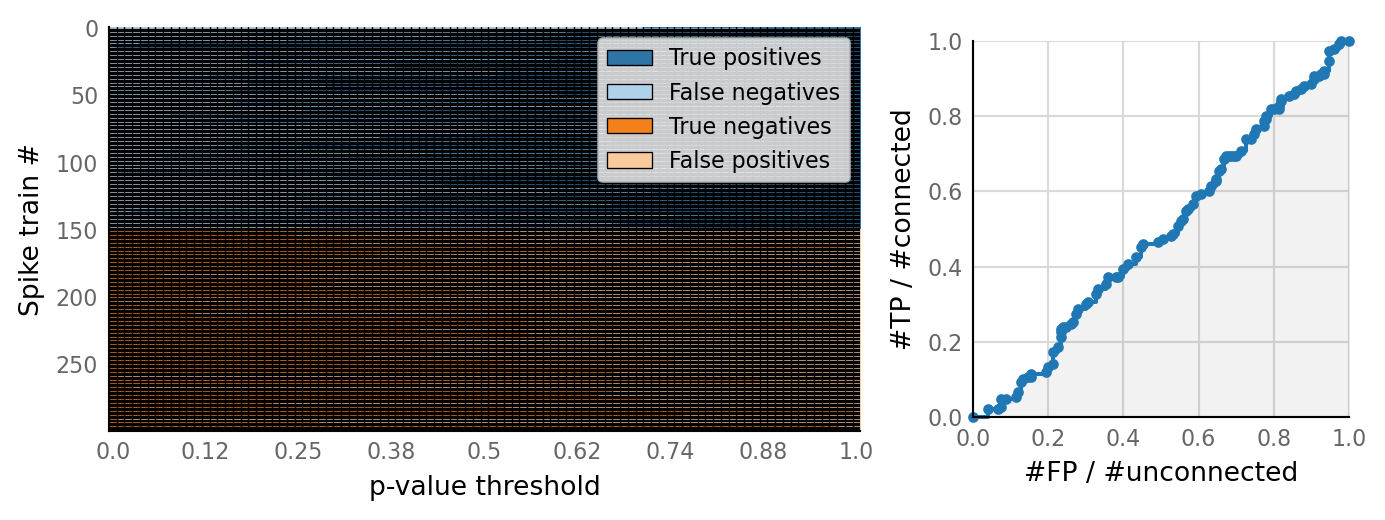
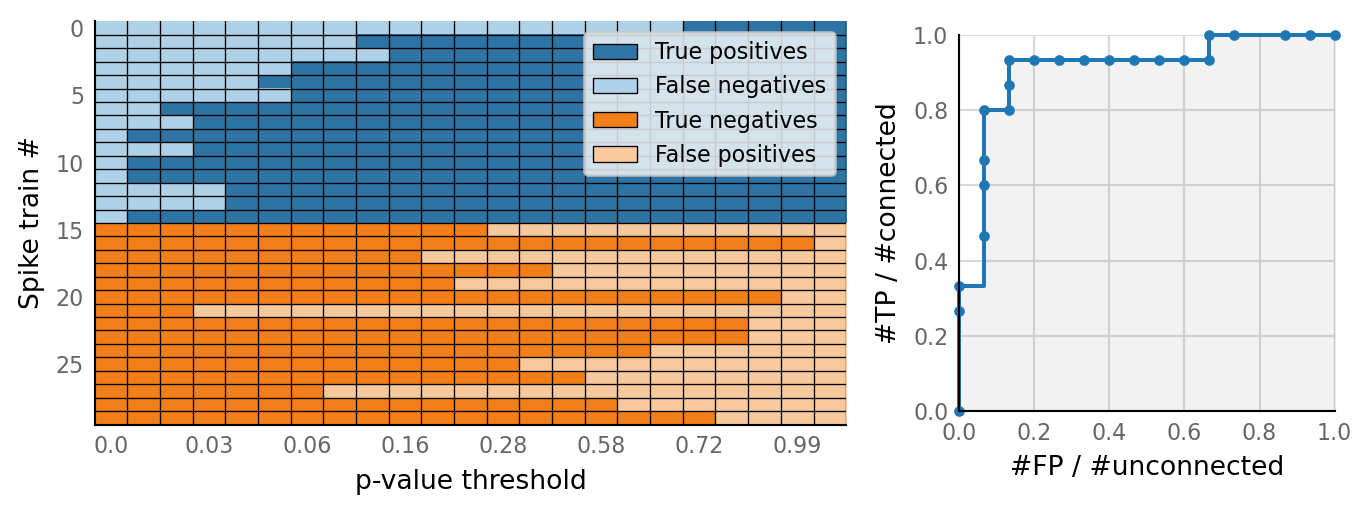
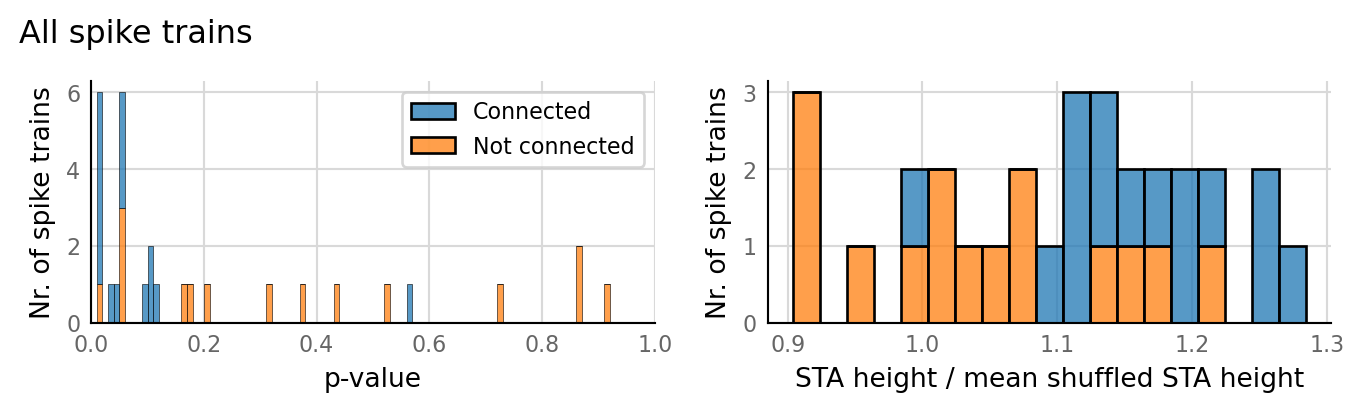
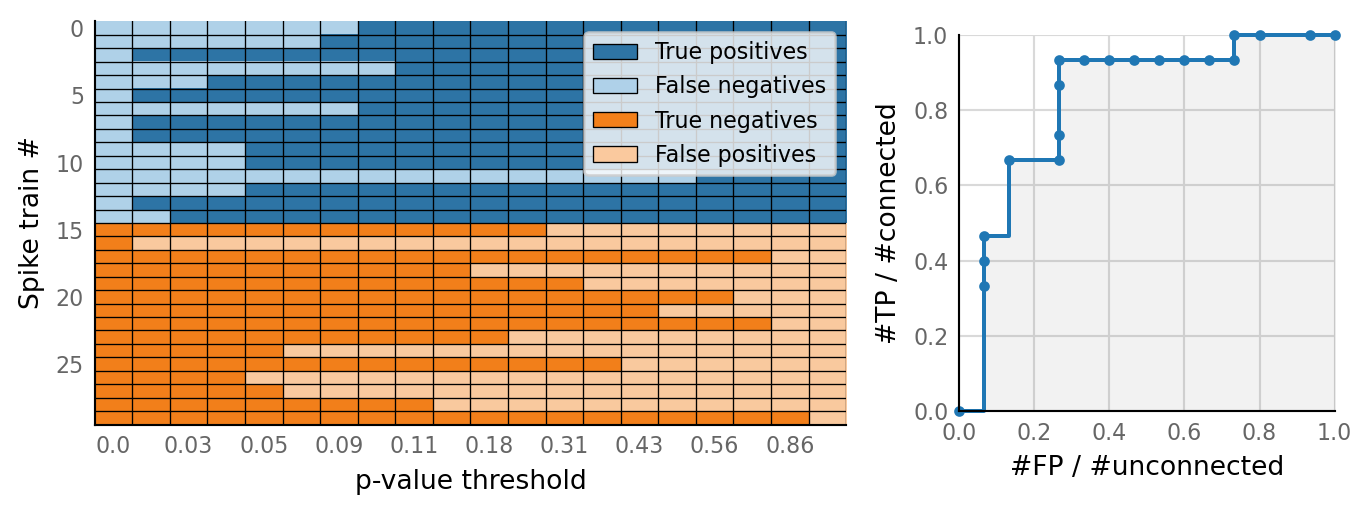
v.N_to_1_experiment.plot_conntest(test_data, test_summaries, sim_data)
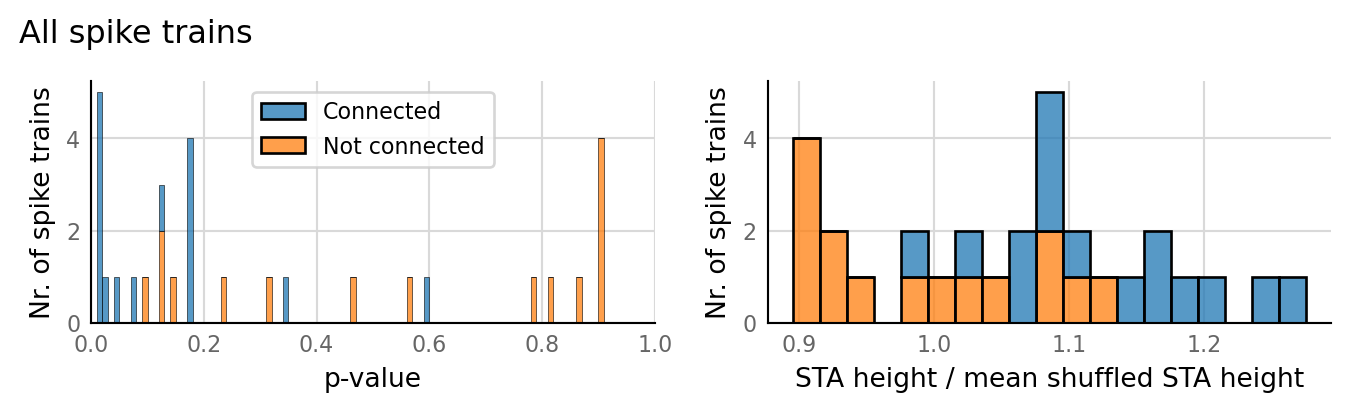
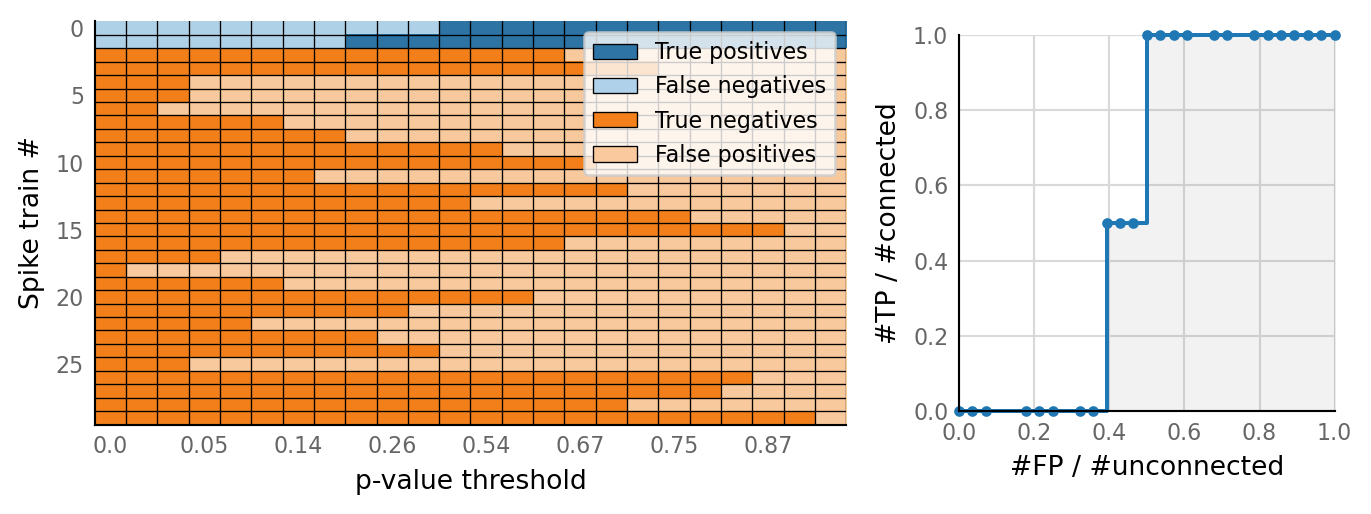
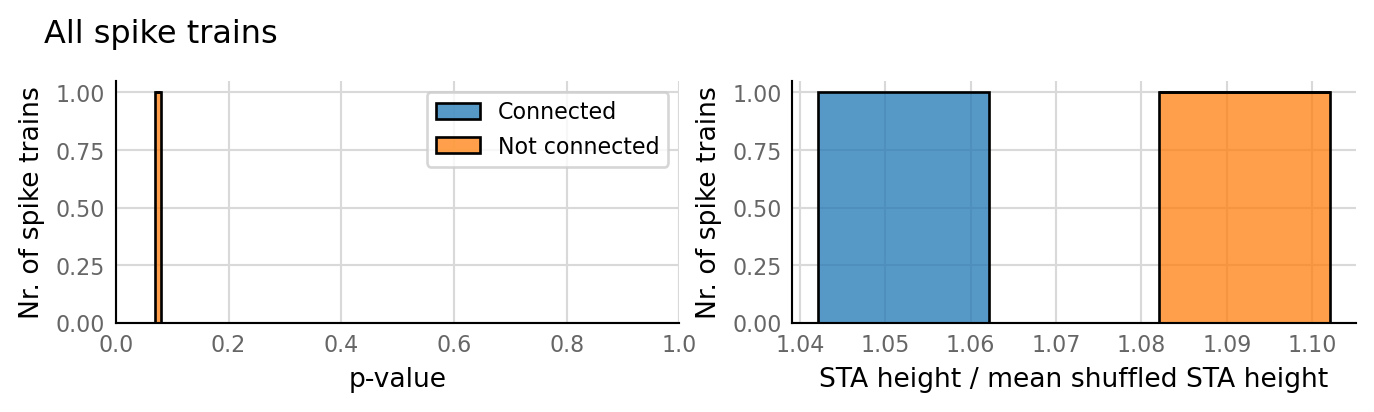
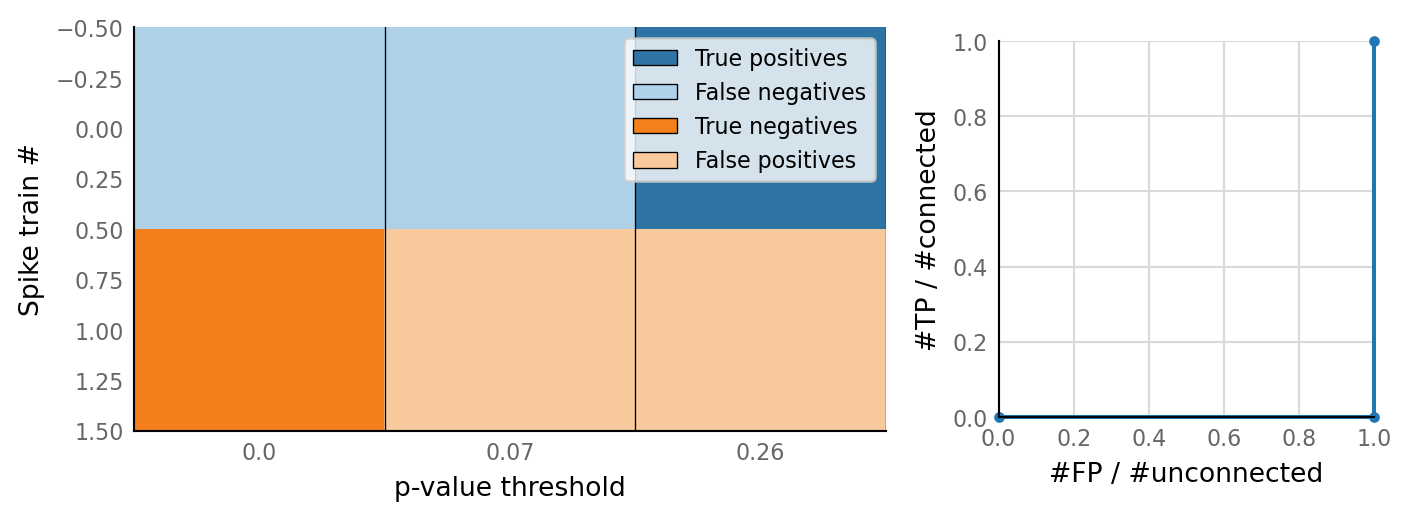
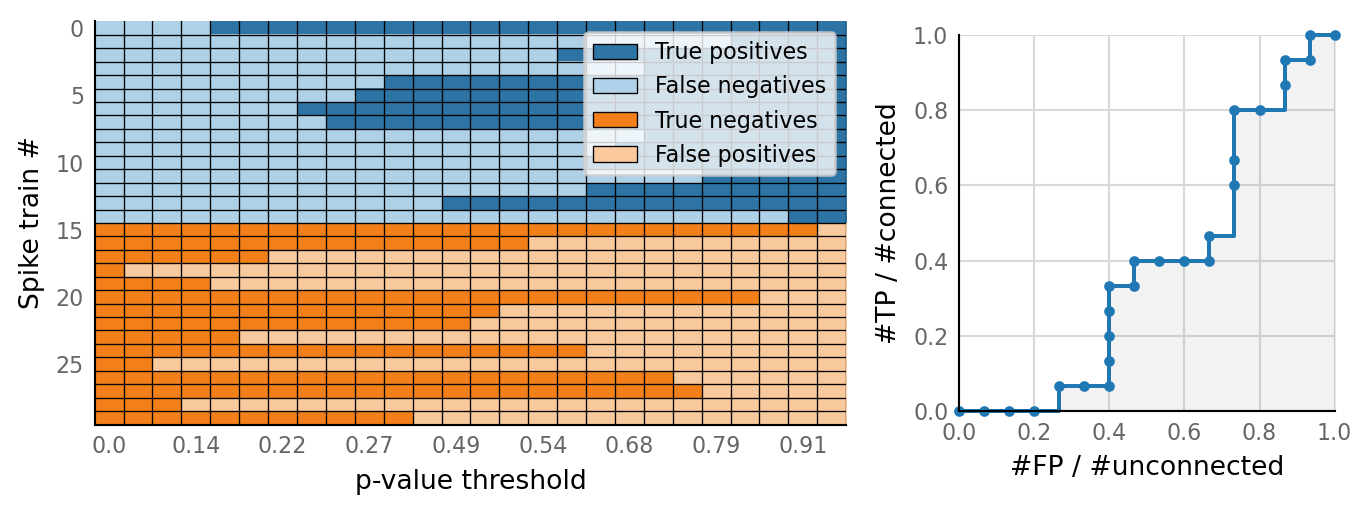
classifs = v.conntest.classification.sweep_threshold(test_summaries, sim_data.is_connected);
v.N_to_1_experiment.plot_classifications_with_ROC(classifs)
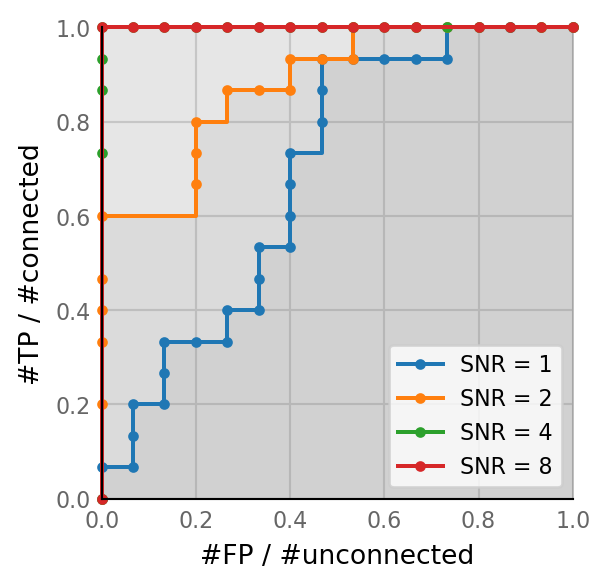
Vary SNR¶
(SNR = voltage imaging signal-to-noise ratio).
Reminder: recording duration is 1 minute, and median reported VI SNR from the literature is 10.
test_connections = v.N_to_1_experiment.test_connections
sweep_threshold = v.conntest.classification.sweep_threshold
def sim_and_eval(params):
with v.time_op("Sim", end=" | "):
sim_data = simulate(params)
with v.time_op("Conntest"):
test_summaries = test_connections(sim_data, inline_meter=True, return_only_summaries=True)
classifs = sweep_threshold(test_summaries, sim_data.is_connected)
return classifs
SNRs = [1, 2, 4, 8];
from copy import deepcopy
%%time
thr_sweeps = []
for SNR in SNRs:
v.bprint(f"SNR: {SNR}", end=" | ")
params = deepcopy(base_params)
params.imaging_spike_SNR = SNR
thr_sweep = sim_and_eval(params)
thr_sweeps.append(thr_sweep)
SNR: 1 | Sim: [0.05 s] | Conntest: [0.04 s]
SNR: 2 | Sim: [0.04 s] | Conntest: [0.04 s]
SNR: 4 | Sim: [0.04 s] | Conntest: [0.04 s]
SNR: 8 | Sim: [0.04 s] | Conntest: [0.04 s]
Wall time: 402 ms
Note: these timings are not real execution time. The results are namely cached, so this is actually time spent finding and reading precomputed results from disk.
plot_ROC = v.conntest.classification.plot_ROC
ax = None
for i, (SNR, classifs) in enumerate(zip(SNRs, thr_sweeps)):
ax = plot_ROC(classifs, ax,
label=f"SNR = {SNR}",
marker='.',
color=f"C{i}")
ax.legend();
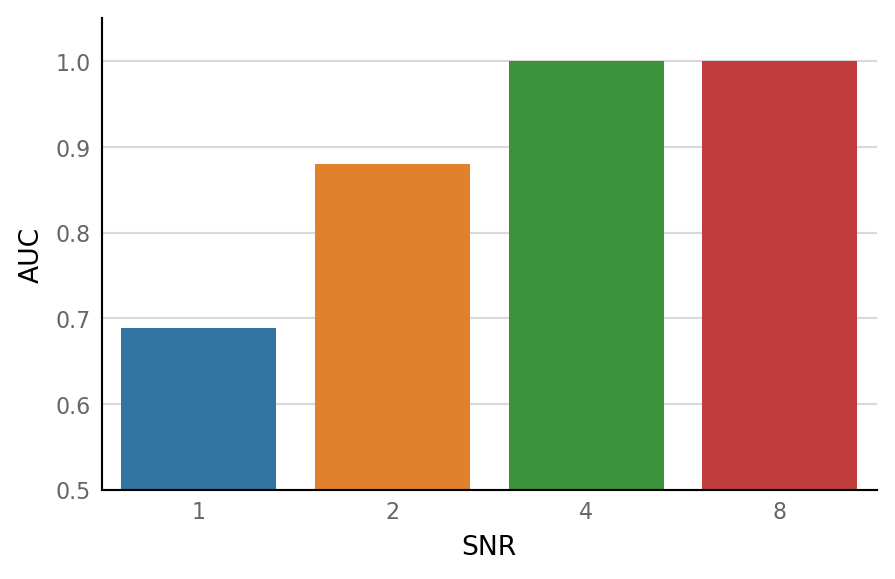
def calc_AUC(thr_sweep):
TPRs = [tr.evaluation.TPR for tr in thr_sweep]
FPRs = [tr.evaluation.FPR for tr in thr_sweep]
AUC = 0
# this is not the same as `np.trapz` e.g.
for i in range(len(FPRs) - 1):
AUC += (FPRs[i+1] - FPRs[i]) * TPRs[i]
return AUC
df = pd.DataFrame(dict(SNR=SNRs, AUC=[calc_AUC(s) for s in thr_sweeps]))
| SNR | AUC | |
|---|---|---|
| 0 | 1 | 0.688889 |
| 1 | 2 | 0.880000 |
| 2 | 4 | 1.000000 |
| 3 | 8 | 1.000000 |
ax = sns.barplot(data=df, x="SNR", y="AUC")
ax.set_ylim(bottom=0.5);
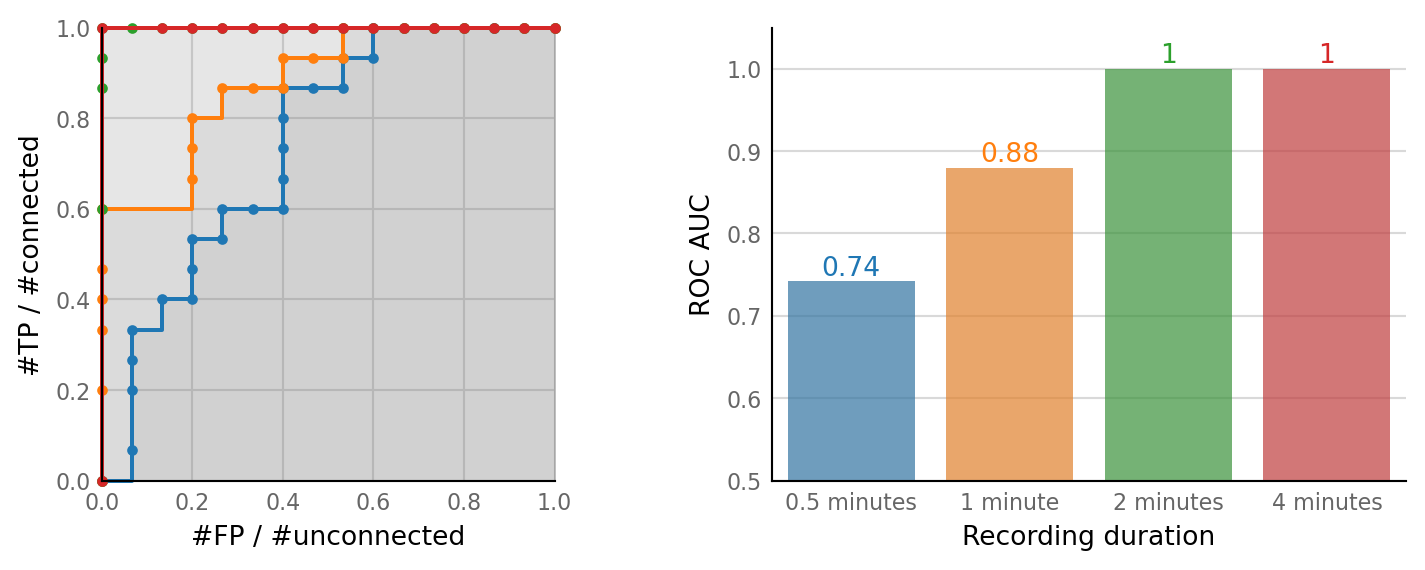
Vary recording duration¶
Reminder: SNR here is 2, and VI photobleaching has time constant of ~10 minutes.
durations = (
30 * second,
1 * minute,
2 * minute,
4 * minute,
);
def format_duration(T):
T_min = T / minute
return f"{T_min:.2G} minute{'' if T_min == 1 else 's'}"
from dataclasses import dataclass, replace
from typing import Any, Callable, TypeVar, Sequence
T = TypeVar('T')
@dataclass
class ParamVariation:
param: str
param_label: str
values: Sequence[T]
value_formatter: Callable[[T], str] = str
def __post_init__(self):
self.paramsets = [replace(base_params, **{self.param: val})
for val in self.values]
self.value_labels = [self.value_formatter(val) for val in self.values]
rec_variation = ParamVariation(
param = "sim_duration",
param_label = "Recording duration",
values = durations,
value_formatter = format_duration,
);
def sim_and_eval_variations(pv: ParamVariation):
max_label_len = max([len(label) for label in pv.value_labels])
thr_sweeps = []
for val, val_label, params in zip(pv.values, pv.value_labels, pv.paramsets):
v.bprint(f"{pv.param_label}: {val_label:<{max_label_len}}", end=" | ")
thr_sweep = sim_and_eval(params)
thr_sweeps.append(thr_sweep)
return thr_sweeps
%time rec_sweeps = sim_and_eval_variations(rec_variation)
Recording duration: 0.5 minutes | Sim: [0.04 s] | Conntest: [0.03 s]
Recording duration: 1 minute | Sim: [0.06 s] | Conntest: [0.04 s]
Recording duration: 2 minutes | Sim: [0.14 s] | Conntest: [0.08 s]
Recording duration: 4 minutes | Sim: [0.35 s] | Conntest: [0.15 s]
Wall time: 946 ms
def barplot_AUC(thr_sweeps, pv: ParamVariation, colors=None, ax=None, text_size=10):
AUCs = [calc_AUC(thr_sweep) for thr_sweep in thr_sweeps]
df = pd.DataFrame({pv.param_label: pv.value_labels, "ROC AUC": AUCs})
ax = v.support.plot_util.new_plot_if_None(ax, **v.figsize(aspect=2.2, width=700))
if colors is None:
colors = ['black'] * len(thr_sweeps)
sns.barplot(data=df, x=pv.param_label, y="ROC AUC", ax=ax, palette=colors, alpha=0.7)
y_bottom = 0.5
ax.set_ylim(bottom=y_bottom)
# Print AUC values as text on bar
for x, AUC, color in zip(ax.get_xticks(), AUCs, colors):
if AUC < y_bottom:
y = y_bottom
else:
y = AUC
ax.text(x, y, format(AUC, ".2G"), va='bottom', ha='center', size=text_size, c=color)
plot_ROC = v.conntest.classification.plot_ROC
def plot_sweeps(thr_sweeps, pv: ParamVariation):
_, (left_ax, right_ax) = plt.subplots(ncols=2, **v.figsize(aspect=3, width=900))
num_sweeps = len(thr_sweeps)
colors = [f"C{i}" for i in range(len(thr_sweeps))]
for sweep, color in zip(thr_sweeps, colors):
plot_ROC(sweep, left_ax, color=color)
barplot_AUC(thr_sweeps, pv, colors, right_ax)
return left_ax, right_ax
left_ax, right_ax = plot_sweeps(rec_sweeps, rec_variation);
Vary p_connected¶
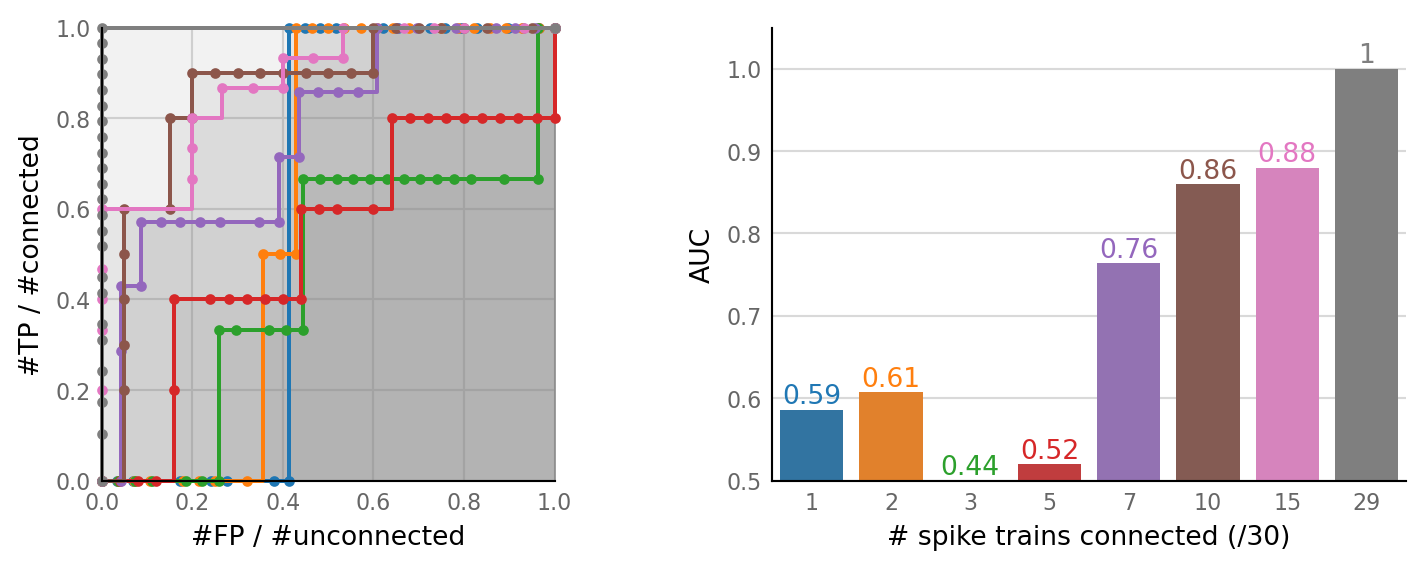
num_connected = (1, 2, 3, 5, 7, 10, 15, 29) # / 30
pconn_variation = ParamVariation(
param = "p_connected",
param_label = "# spike trains connected (/30)",
values = np.array(num_connected) / 30,
value_formatter=lambda val: str(int(val * 30)),
);
%time pconn_sweeps = sim_and_eval_variations(pconn_variation)
# spike trains connected (/30): 1 | Sim: [0.10 s] | Conntest: [0.05 s]
# spike trains connected (/30): 2 | Sim: [0.08 s] | Conntest: [0.05 s]
# spike trains connected (/30): 3 | Sim: [0.08 s] | Conntest: [0.05 s]
# spike trains connected (/30): 5 | Sim: [0.08 s] | Conntest: [0.05 s]
# spike trains connected (/30): 7 | Sim: [0.08 s] | Conntest: [0.05 s]
# spike trains connected (/30): 10 | Sim: [0.07 s] | Conntest: [0.05 s]
# spike trains connected (/30): 15 | Sim: [0.08 s] | Conntest: [0.05 s]
# spike trains connected (/30): 29 | Sim: [0.08 s] | Conntest: [0.05 s]
Wall time: 1.09 s
plot_sweeps(pconn_sweeps, pconn_variation);
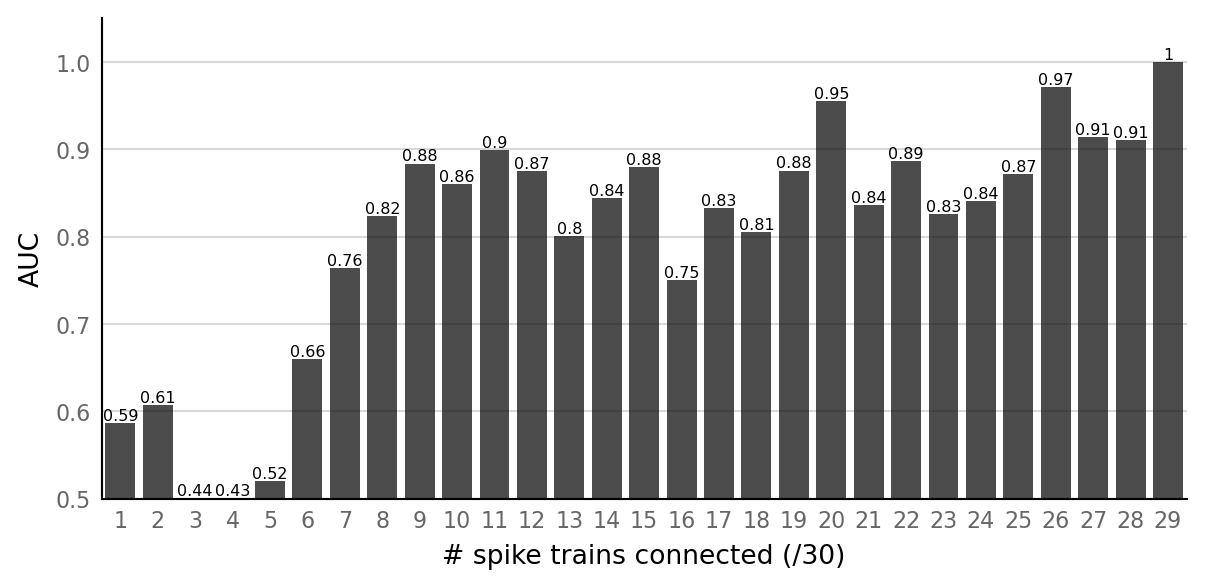
pconn_variation_full = replace(pconn_variation, values = np.arange(1, 30) / 30);
(We don’t try 0 connected, as then TPR becomes meaningless: there are no positives and thus no true positives.
Same for all 30 connected, where FPR becomes meaningless: there are no negatives and thus no false positives).
%%time
pconn_sweeps_full = sim_and_eval_variations(pconn_variation_full)
# spike trains connected (/30): 1 | Sim: [0.04 s] | Conntest: [0.04 s]
# spike trains connected (/30): 2 | Sim: [0.04 s] | Conntest: [0.04 s]
# spike trains connected (/30): 3 | Sim: [0.04 s] | Conntest: [0.04 s]
# spike trains connected (/30): 4 | Sim: [0.04 s] | Conntest: [0.04 s]
# spike trains connected (/30): 5 | Sim: [0.03 s] | Conntest: [0.04 s]
# spike trains connected (/30): 6 | Sim: [0.03 s] | Conntest: [0.04 s]
# spike trains connected (/30): 7 | Sim: [0.03 s] | Conntest: [0.04 s]
# spike trains connected (/30): 8 | Sim: [0.03 s] | Conntest: [0.04 s]
# spike trains connected (/30): 9 | Sim: [0.03 s] | Conntest: [0.04 s]
# spike trains connected (/30): 10 | Sim: [0.03 s] | Conntest: [0.04 s]
# spike trains connected (/30): 11 | Sim: [0.03 s] | Conntest: [0.04 s]
# spike trains connected (/30): 12 | Sim: [0.05 s] | Conntest: [0.05 s]
# spike trains connected (/30): 13 | Sim: [0.04 s] | Conntest: [0.04 s]
# spike trains connected (/30): 14 | Sim: [0.04 s] | Conntest: [0.04 s]
# spike trains connected (/30): 15 | Sim: [0.04 s] | Conntest: [0.04 s]
# spike trains connected (/30): 16 | Sim: [0.04 s] | Conntest: [0.04 s]
# spike trains connected (/30): 17 | Sim: [0.04 s] | Conntest: [0.04 s]
# spike trains connected (/30): 18 | Sim: [0.04 s] | Conntest: [0.04 s]
# spike trains connected (/30): 19 | Sim: [0.04 s] | Conntest: [0.04 s]
# spike trains connected (/30): 20 | Sim: [0.03 s] | Conntest: [0.04 s]
# spike trains connected (/30): 21 | Sim: [0.04 s] | Conntest: [0.04 s]
# spike trains connected (/30): 22 | Sim: [0.05 s] | Conntest: [0.04 s]
# spike trains connected (/30): 23 | Sim: [0.04 s] | Conntest: [0.04 s]
# spike trains connected (/30): 24 | Sim: [0.04 s] | Conntest: [0.04 s]
# spike trains connected (/30): 25 | Sim: [0.04 s] | Conntest: [0.06 s]
# spike trains connected (/30): 26 | Sim: [0.05 s] | Conntest: [0.05 s]
# spike trains connected (/30): 27 | Sim: [0.03 s] | Conntest: [0.04 s]
# spike trains connected (/30): 28 | Sim: [0.03 s] | Conntest: [0.04 s]
# spike trains connected (/30): 29 | Sim: [0.03 s] | Conntest: [0.04 s]
Wall time: 2.62 s
(Running this the first time, with 19 of the 29 param values not run yet – i.e. only 10 where loaded from disk cache – took 44 seconds).
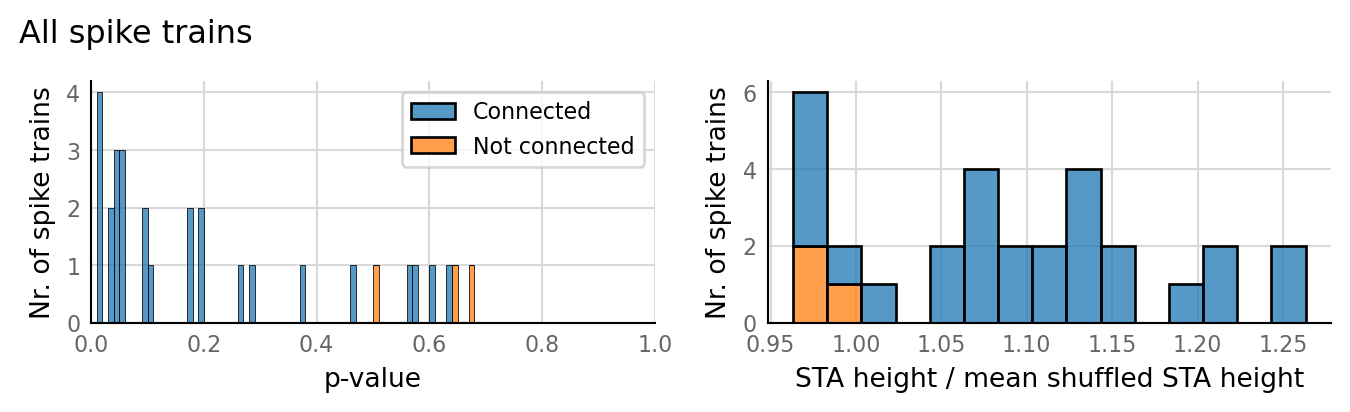
barplot_AUC(pconn_sweeps_full, pconn_variation_full, text_size=6)
Inspect some simulations¶
show_output_spike_stats = v.sim.izhikevich_neuron.show_output_spike_stats
plot_conntest = v.N_to_1_experiment.plot_conntest
plot_classifications_with_ROC = v.N_to_1_experiment.plot_classifications_with_ROC
def sim_eval_plot(params):
with v.time_op("Sim"):
sim_data = simulate(params)
plot_sim_slice(sim_data, t_start=3 * second, duration=1 * second)
show_output_spike_stats(sim_data.izh_output)
with v.time_op("Conntest"):
test_data, test_summaries = test_connections(sim_data, inline_meter=True)
plot_conntest(test_data, test_summaries, sim_data)
classifs = sweep_threshold(test_summaries, sim_data.is_connected)
plot_classifications_with_ROC(classifs)
return sim_data, test_data, test_summaries, classifs
2 spike trains connected:¶
No output spikes.
sim_eval_plot(pconn_variation_full.paramsets[1]);
Sim: [0.03 s]
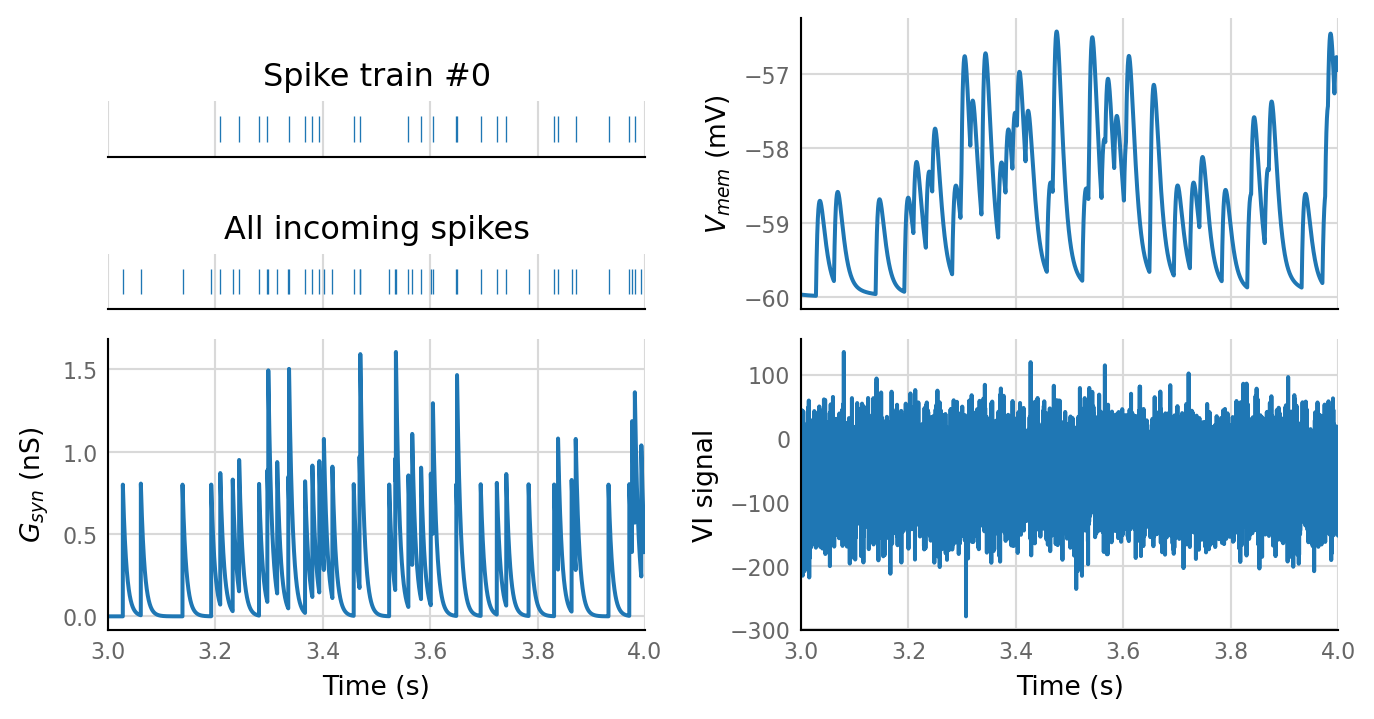
Output spike rate (1 / median ISI): NAN Hz
Conntest:
C:\conda\lib\site-packages\numpy\core\fromnumeric.py:3372: RuntimeWarning: Mean of empty slice.
return _methods._mean(a, axis=axis, dtype=dtype,
C:\conda\lib\site-packages\numpy\core\_methods.py:170: RuntimeWarning: invalid value encountered in double_scalars
ret = ret.dtype.type(ret / rcount)
[0.77 s]
We could make a better detector by looking at the slope of the first ~20 ms of the STA, somehow denoised.
Or maybe a local median filter over the STA, then still take ptp (max-min) ?
6 spike trains connected:¶
sim_eval_plot(pconn_variation_full.paramsets[5]);
Sim: [0.03 s]
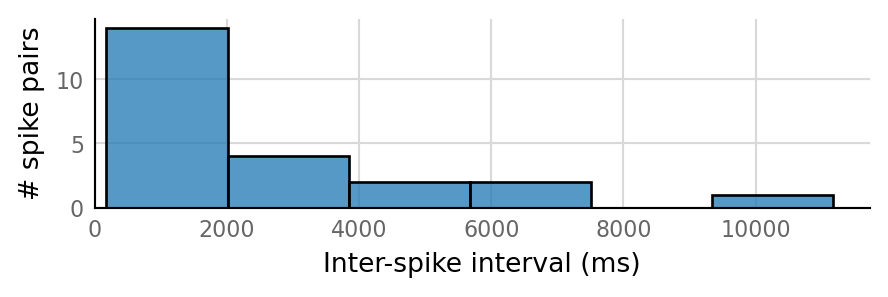
Output spike rate (1 / median ISI): 0.553 Hz
Conntest: [0.42 s]
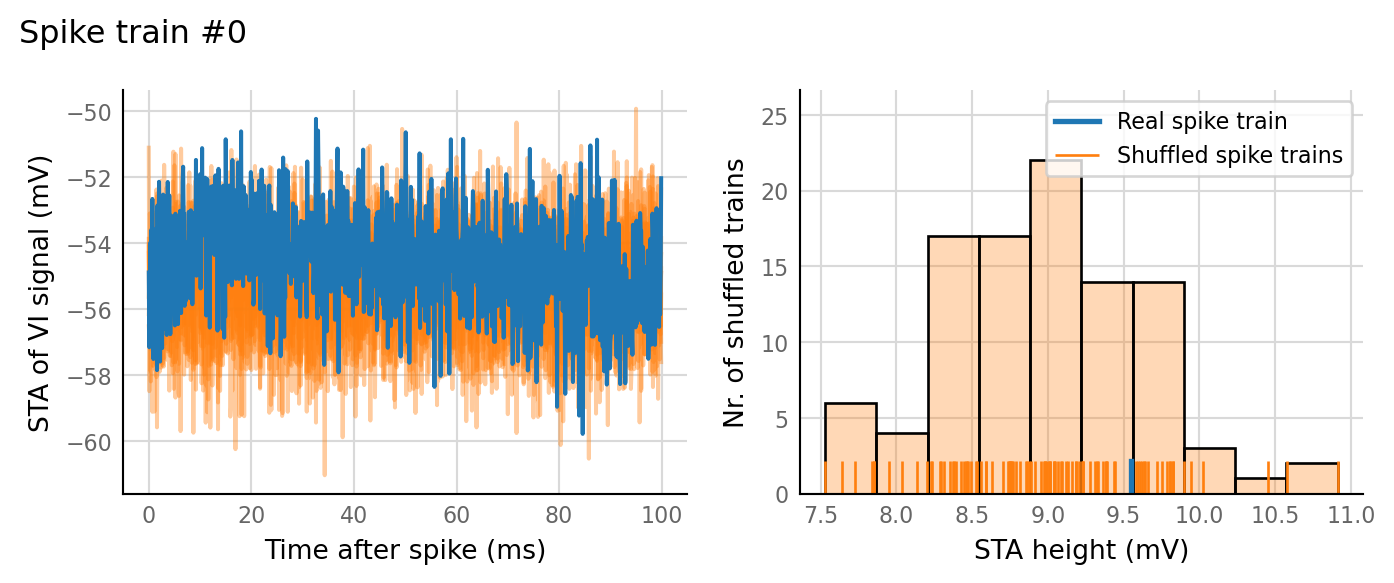
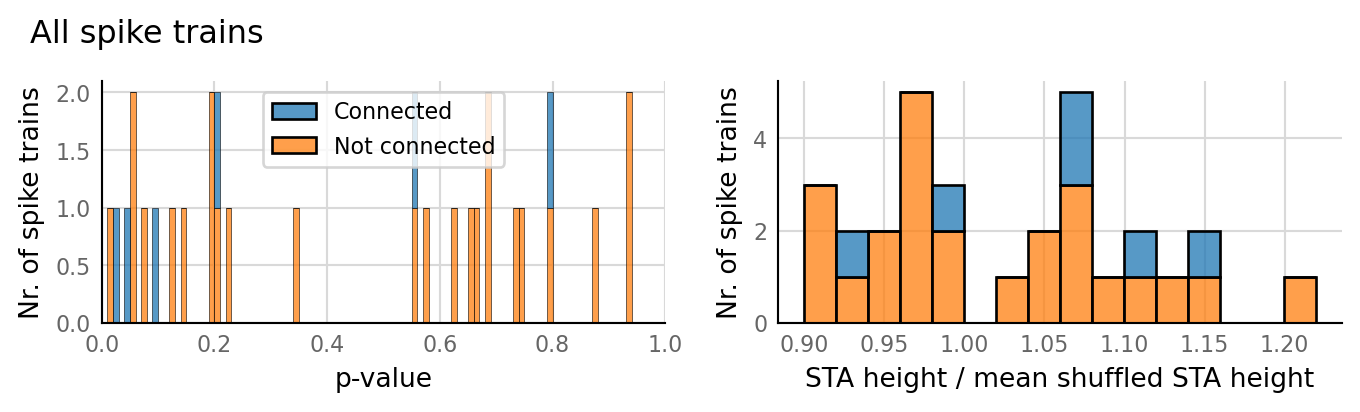
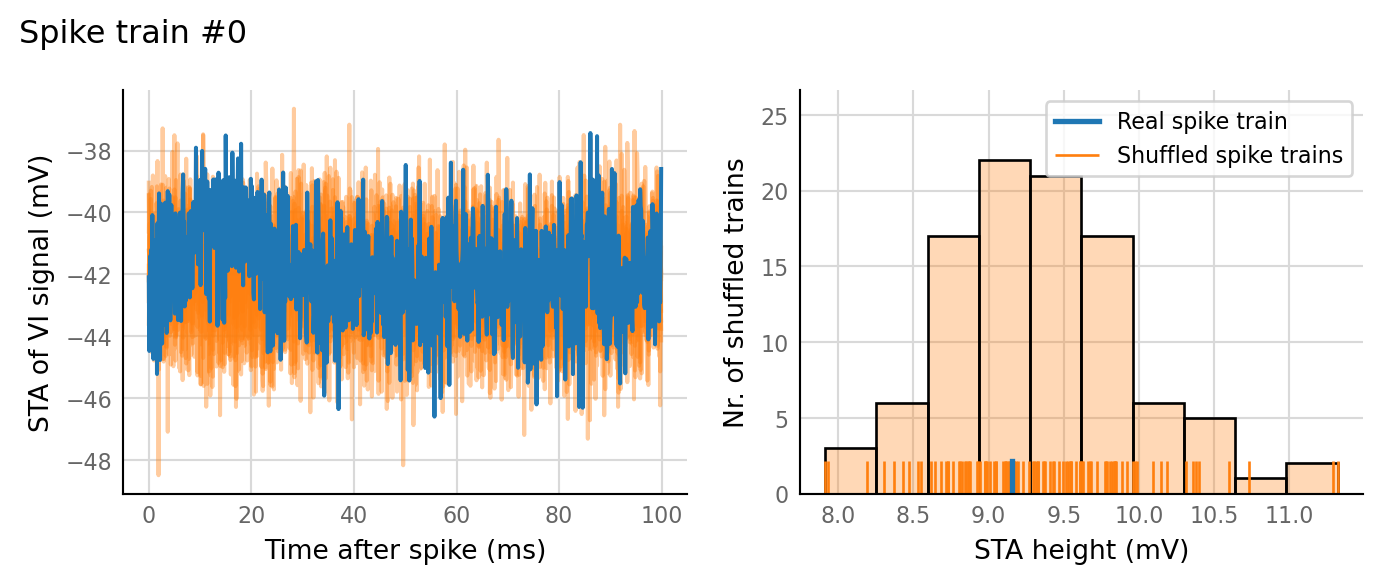
27 spike trains connected:¶
sim_eval_plot(pconn_variation_full.paramsets[26]);
Sim: [0.03 s]
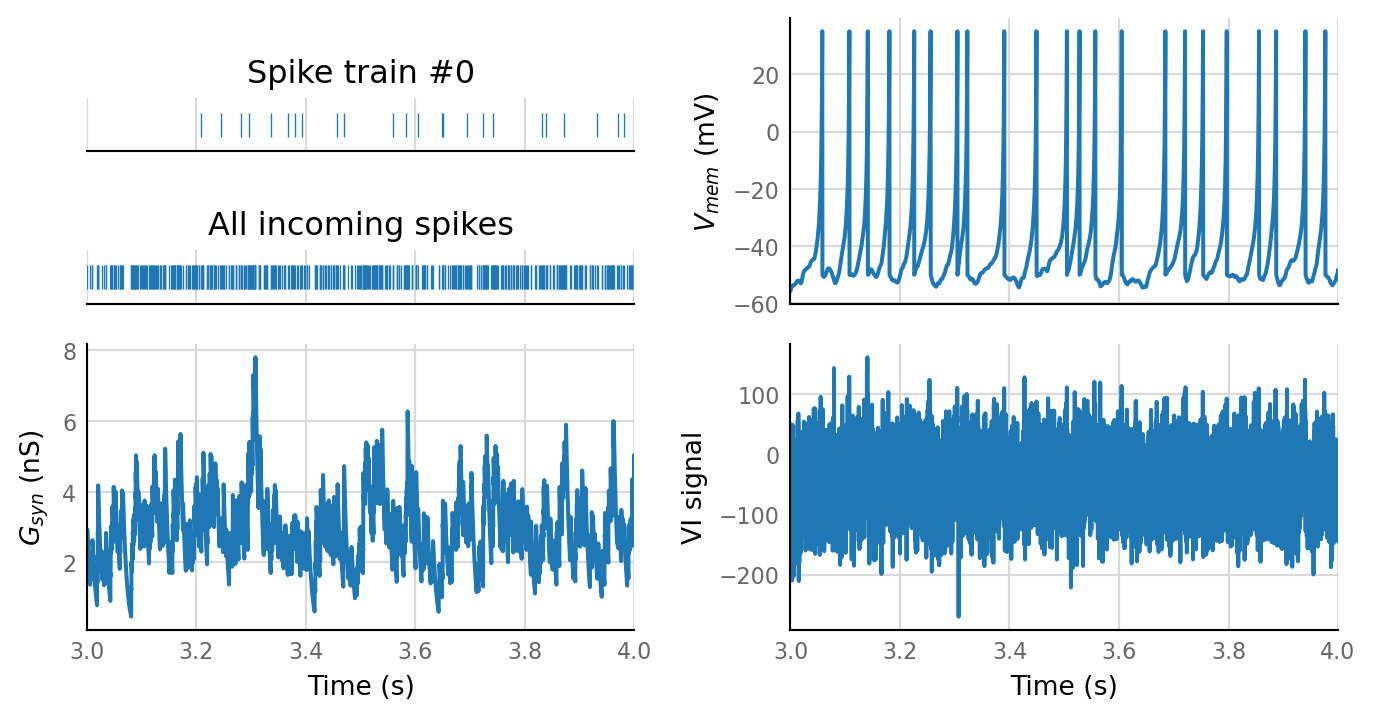
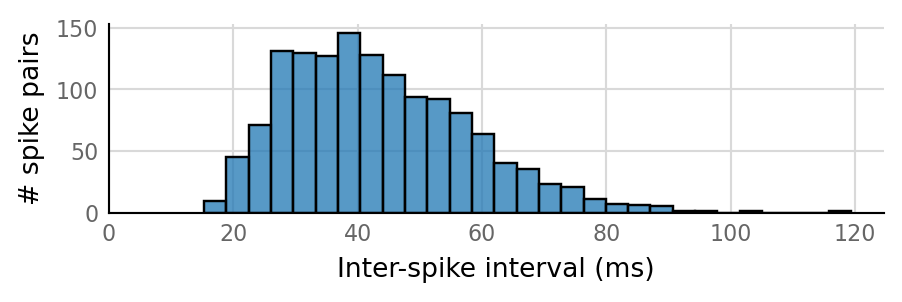
Output spike rate (1 / median ISI): 24.2 Hz
Conntest: [0.41 s]
Question¶
Why do the simulations with low p_connected have low AUC? There are still EPSPs that can be averaged.
The increase in AUC with increasing p_connected suspiciously coincides with the appearance of output spikes.
So is STA bump due to spikes after all?
Weird cause I found 2020-10-23 • Delete signal around spikes before STA sufficiently convincing: deleting 60 ms before each output spike to 20 ms after.
That notebook did have 10 minute recording duration & SNR 10 though (here: 1 minute & SNR 2).
Vary # spike trains¶
..half of which are connected.
N_variation = ParamVariation(
param = "num_spike_trains",
param_label = "# spike trains",
values = (2, 16, 30, 60, 300),
value_formatter=lambda val: str(int(val)),
);
%time N_sweeps = sim_and_eval_variations(N_variation)
# spike trains: 2 | Sim: [0.03 s] | Conntest: [0.04 s]
# spike trains: 16 | Sim: [0.03 s] | Conntest: [0.04 s]
# spike trains: 30 | Sim: [0.03 s] | Conntest: [0.04 s]
# spike trains: 60 | Sim: [0.03 s] | Conntest: [0.05 s]
# spike trains: 300 | Sim: [0.05 s] | Conntest: [0.07 s]
Wall time: 454 ms
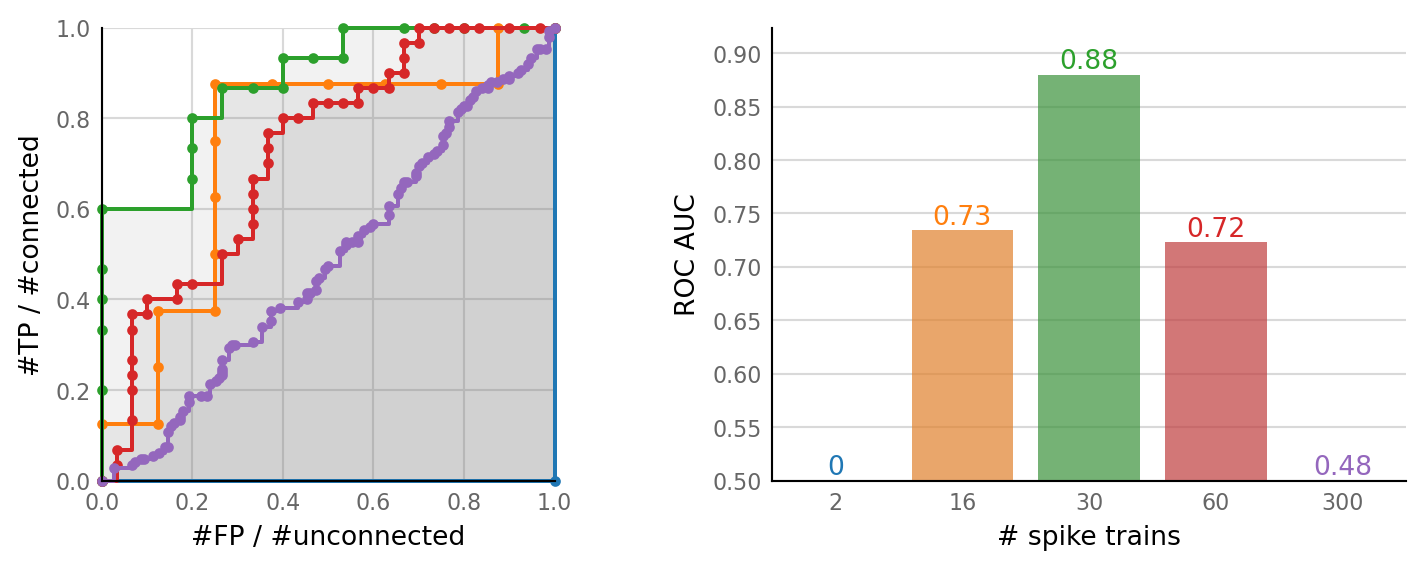
plot_sweeps(N_sweeps, N_variation);
Inspect some simulations¶
sim_eval_plot(N_variation.paramsets[0]);
Sim: [0.04 s]
Output spike rate (1 / median ISI): NAN Hz
Conntest: [0.07 s]
C:\conda\lib\site-packages\numpy\core\fromnumeric.py:3372: RuntimeWarning: Mean of empty slice.
return _methods._mean(a, axis=axis, dtype=dtype,
C:\conda\lib\site-packages\numpy\core\_methods.py:170: RuntimeWarning: invalid value encountered in double_scalars
ret = ret.dtype.type(ret / rcount)
sim_eval_plot(N_variation.paramsets[1]);
Sim: [0.04 s]
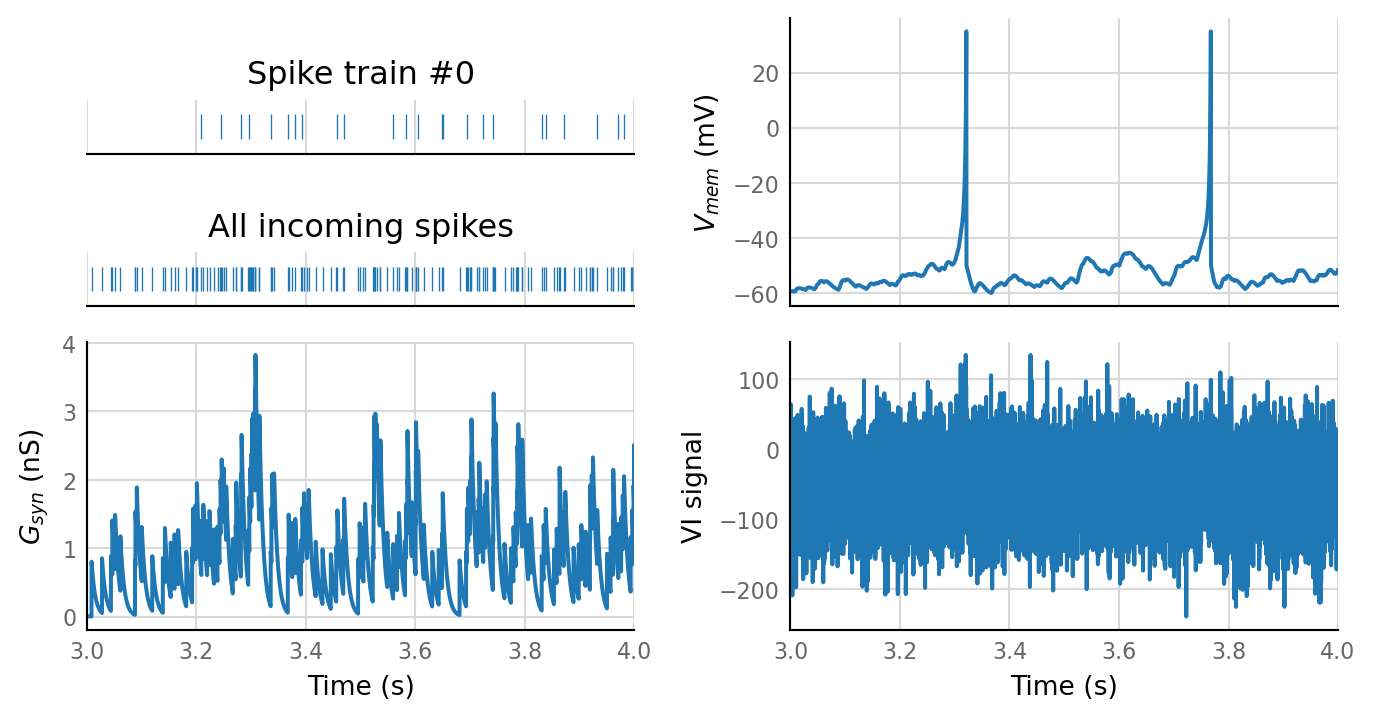
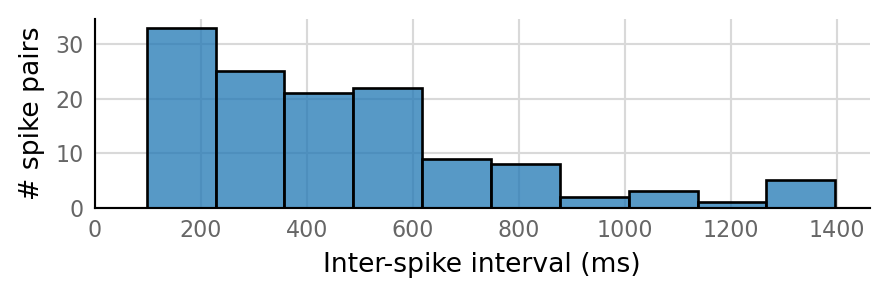
Output spike rate (1 / median ISI): 2.58 Hz
Conntest: [0.29 s]
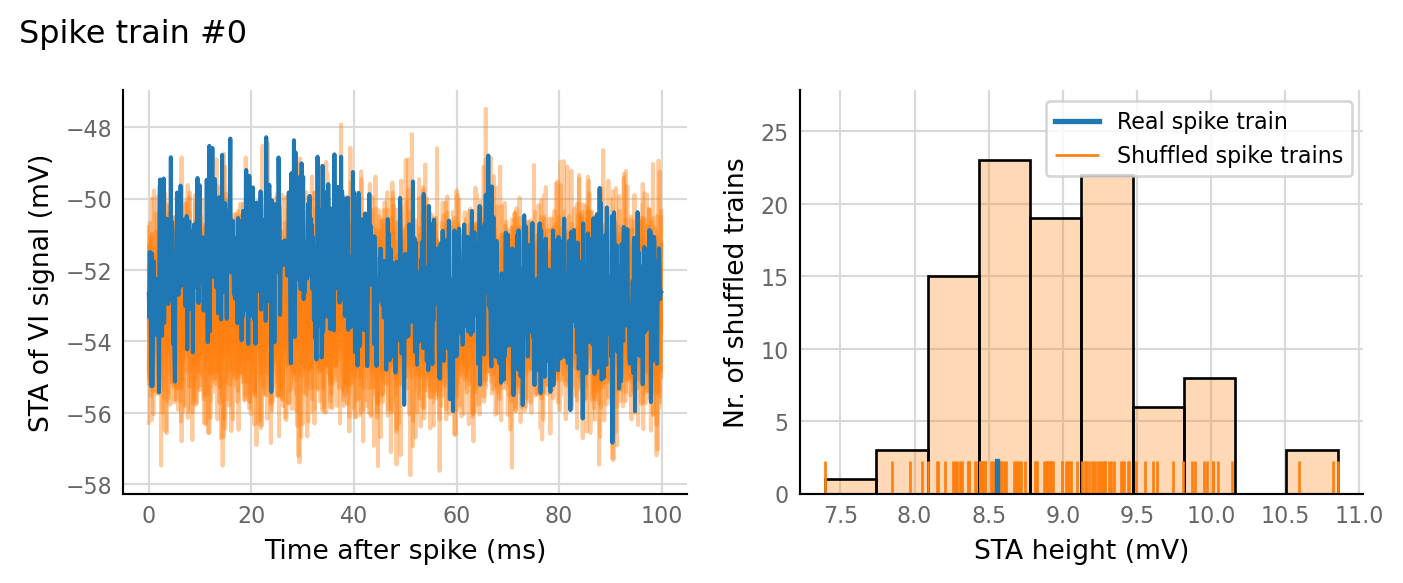
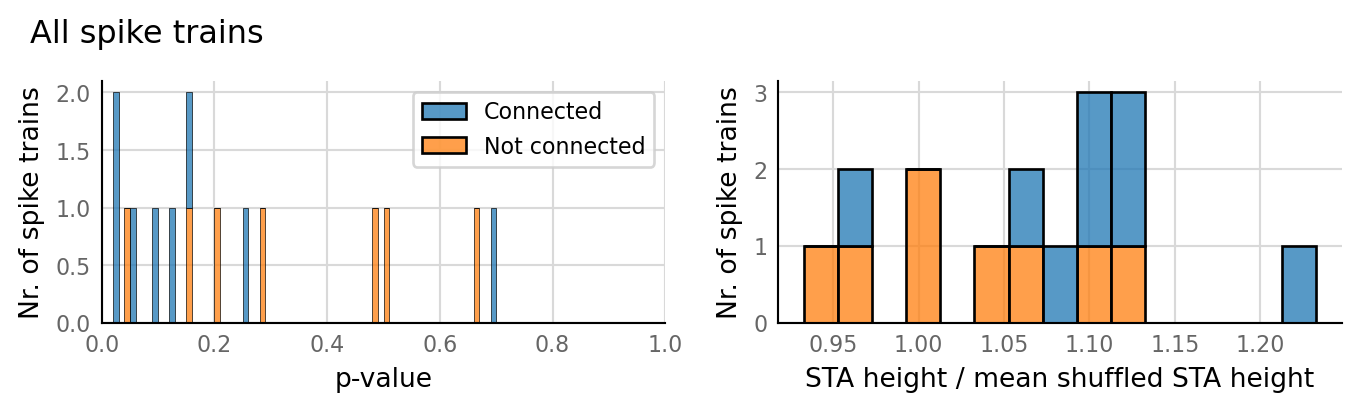
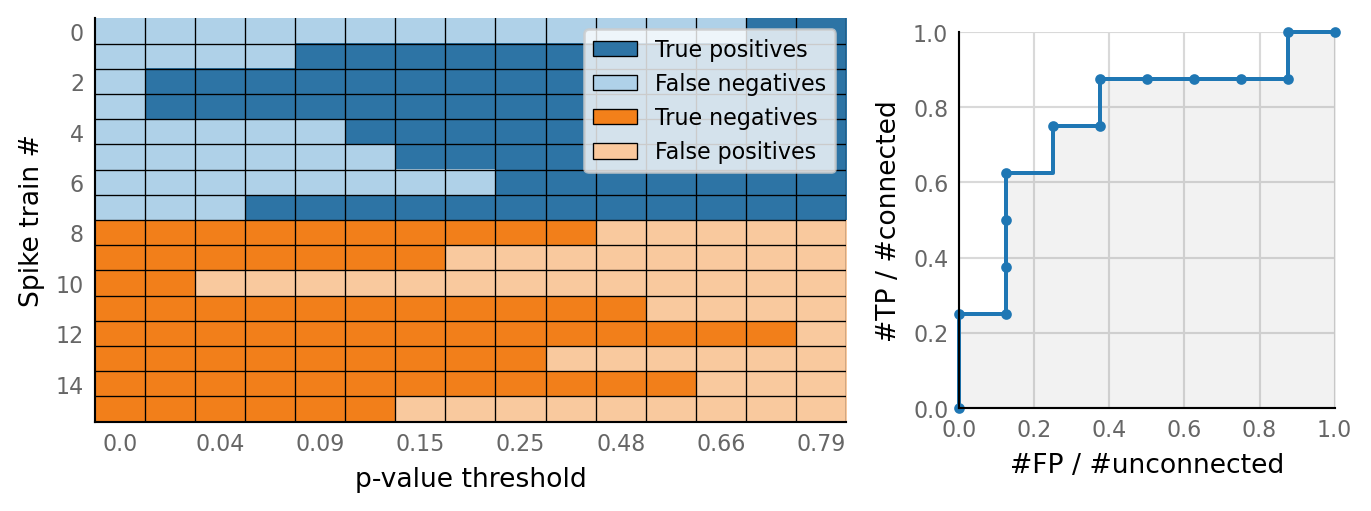
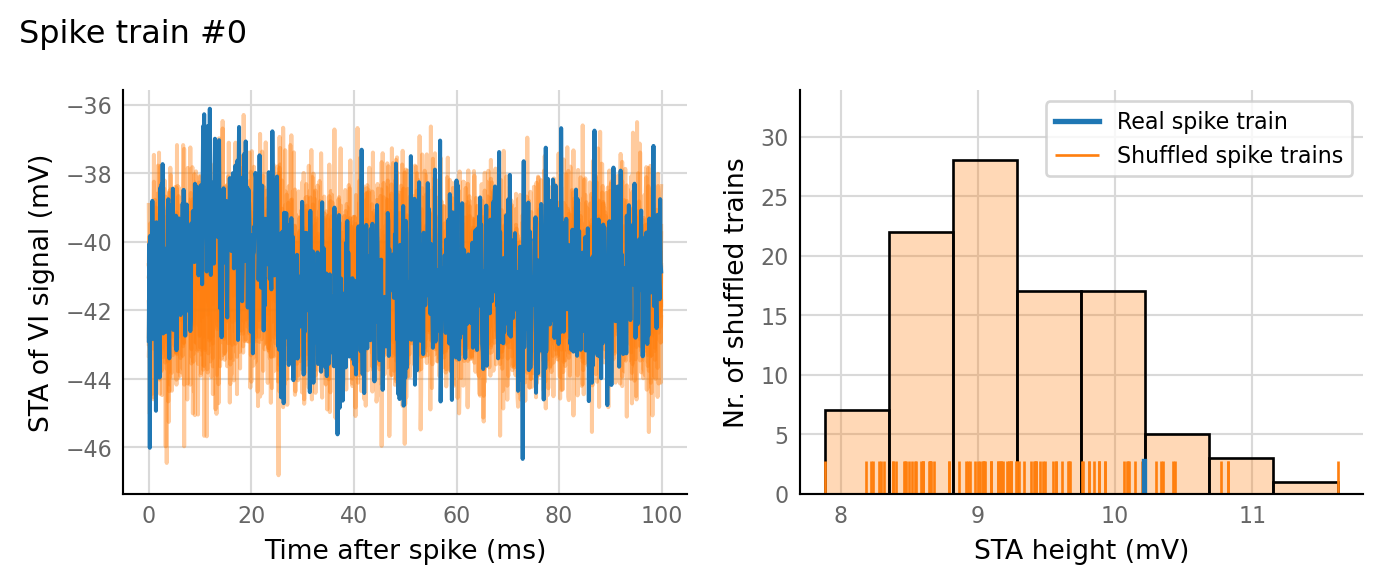
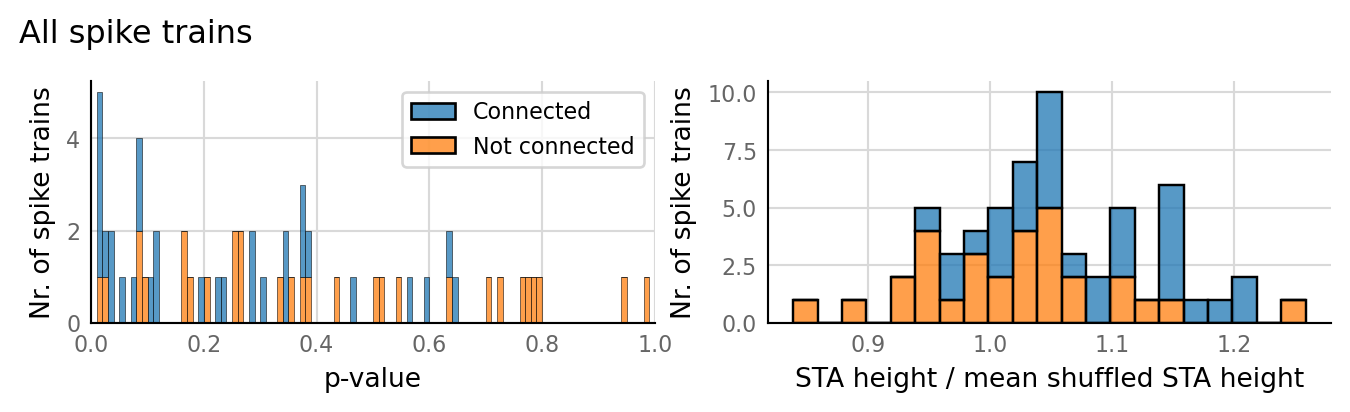
sim_eval_plot(N_variation.paramsets[3]);
Sim: [0.04 s]
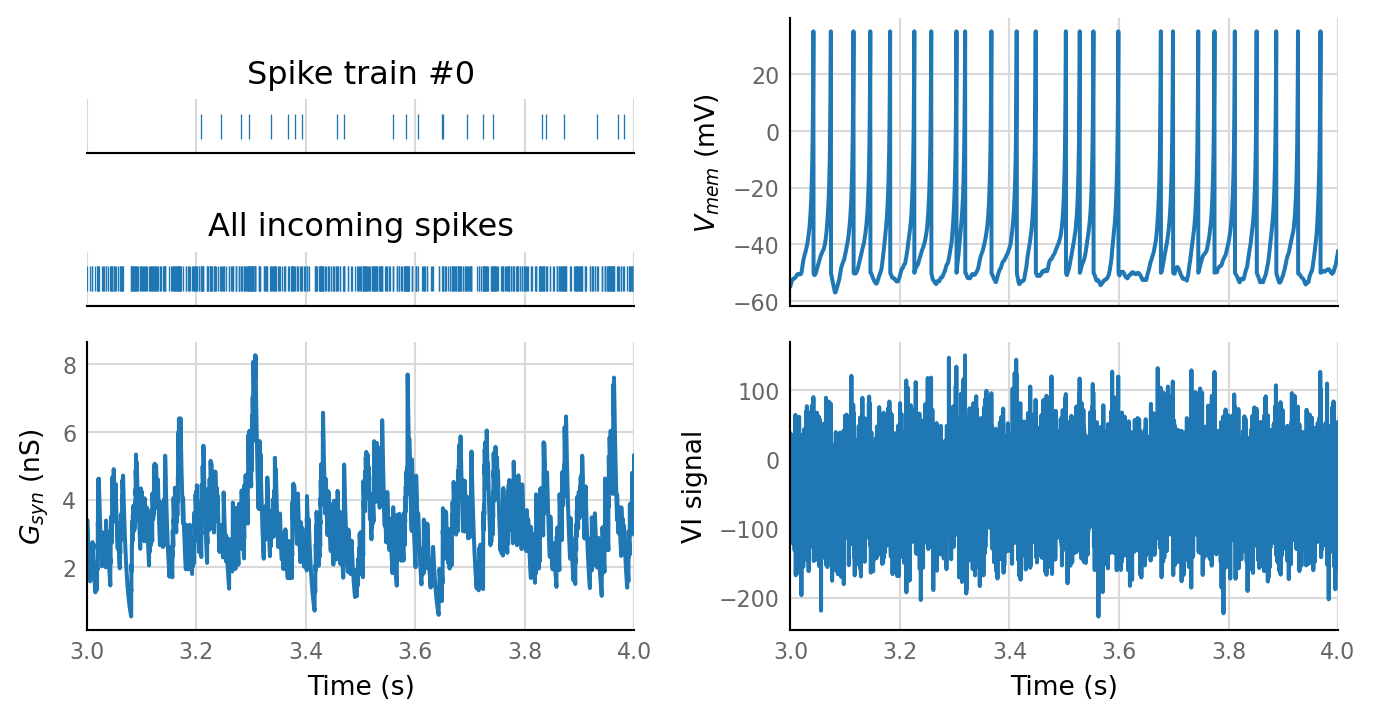
Output spike rate (1 / median ISI): 27.1 Hz
Conntest: [1.13 s]
sim_eval_plot(N_variation.paramsets[4]);
Sim: [0.06 s]
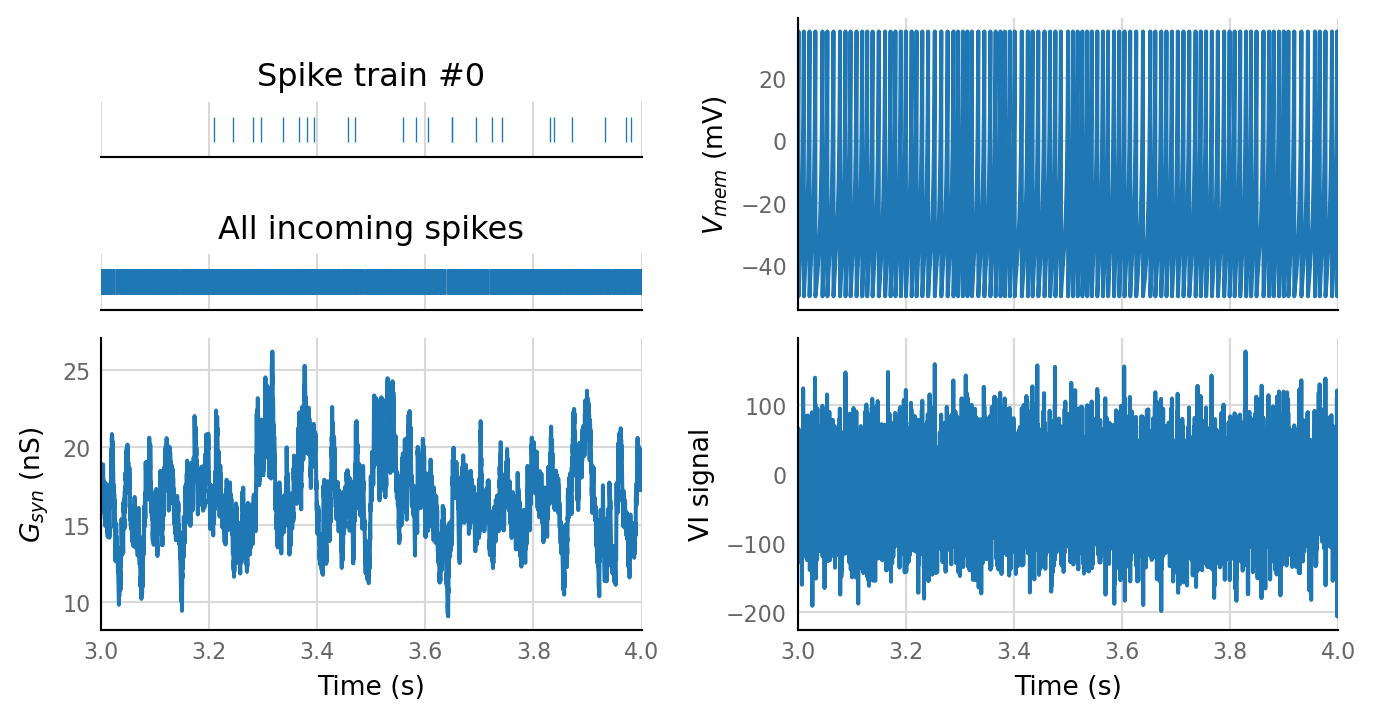
Output spike rate (1 / median ISI): 98 Hz
Conntest: [4.67 s]
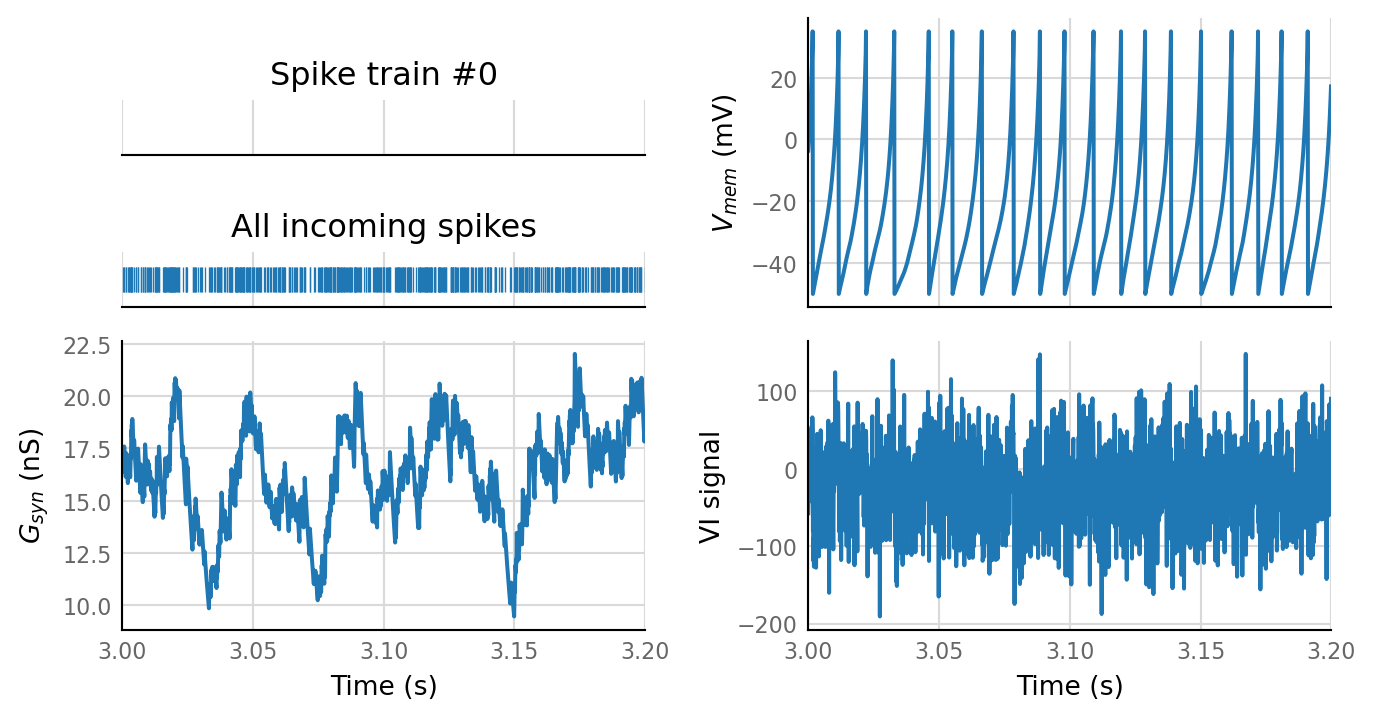
^ 300 incoming neurons. Spike rate does seem to follow synaptic conductance.
Zoom in:
sd = simulate(N_variation.paramsets[4])
plot_sim_slice(sd, t_start=3*second, duration=0.2 * second);
Discussion¶
For these parameters, we thus get best detection performance @ output spike rate ≈ 11 Hz – what we get for N = 30 spike trains here.
Output spike rates of 2.6 Hz (N = 16) and 27 Hz (N = 60) are about equally worse.
Vary \(\tau_{syn}\)¶
base_params
N_to_1_SimParams(sim_duration=60, timestep=0.0001, num_spike_trains=30, p_connected=0.5, spike_rate=20.0, Δg_syn=8.000000000000001e-10, τ_syn=0.007, neuron_params=IzhikevichParams(C=1e-10, k=7e-07, v_r=-0.06, v_t=-0.04, v_peak=0.035, a=30.0, b=-2e-09, c=-0.05, d=1e-10, v_syn=0.0), imaging_spike_SNR=2, rng_seed=1800)
tau_variation = ParamVariation(
param = "τ_syn",
param_label = r"$\tau_{syn}$",
values = np.array([1, 3, 7, 14, 70]) * ms,
value_formatter=lambda val: f"{int(val / ms)} ms",
);
%time tau_sweeps = sim_and_eval_variations(tau_variation)
$\tau_{syn}$: 1 ms | Sim: [0.03 s] | Conntest: [0.04 s]
$\tau_{syn}$: 3 ms | Sim: [0.03 s] | Conntest: [0.04 s]
$\tau_{syn}$: 7 ms | Sim: [0.03 s] | Conntest: [0.04 s]
$\tau_{syn}$: 14 ms | Sim: [0.03 s] | Conntest: [0.04 s]
$\tau_{syn}$: 70 ms | Sim: [0.03 s] | Conntest: [0.04 s]
Wall time: 419 ms
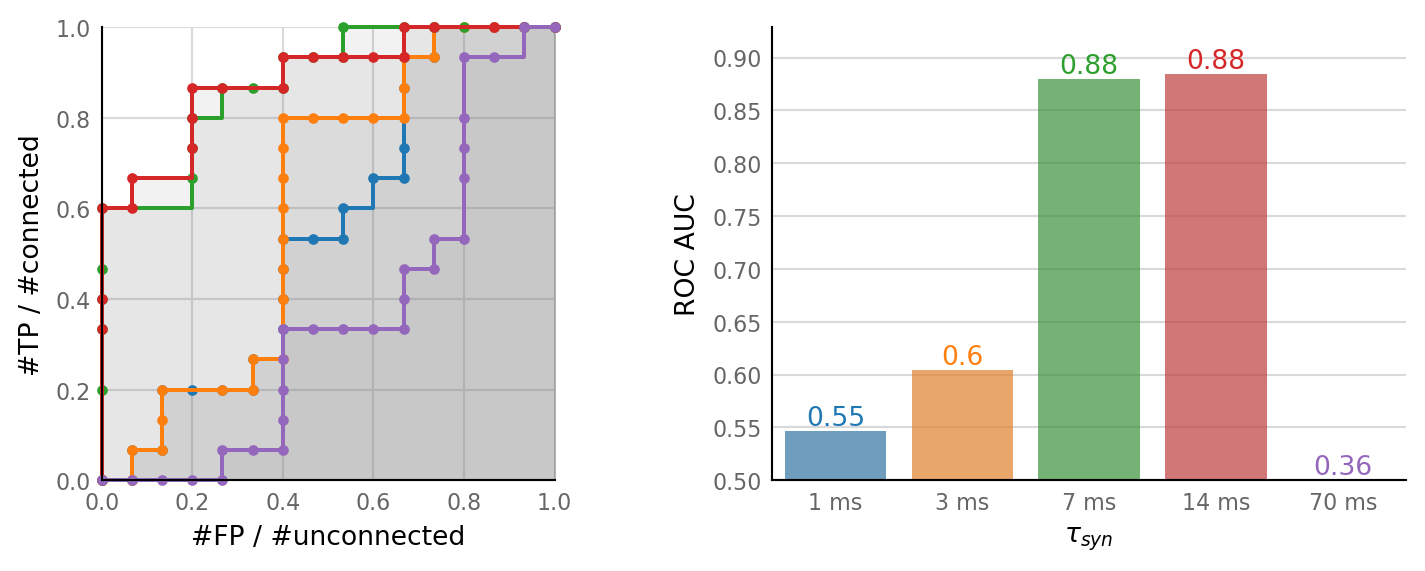
plot_sweeps(tau_sweeps, tau_variation);
Inspect some simulations¶
sim_eval_plot(tau_variation.paramsets[1]);
Sim: [0.04 s]
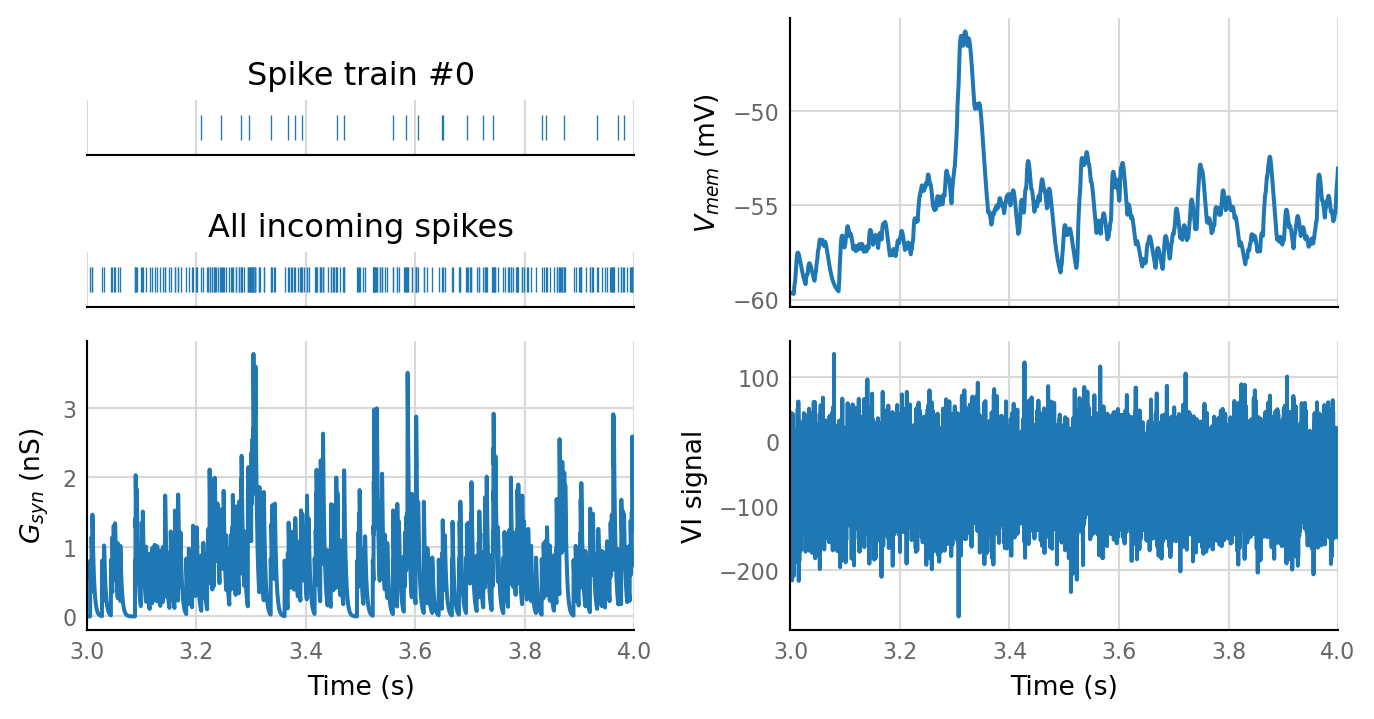
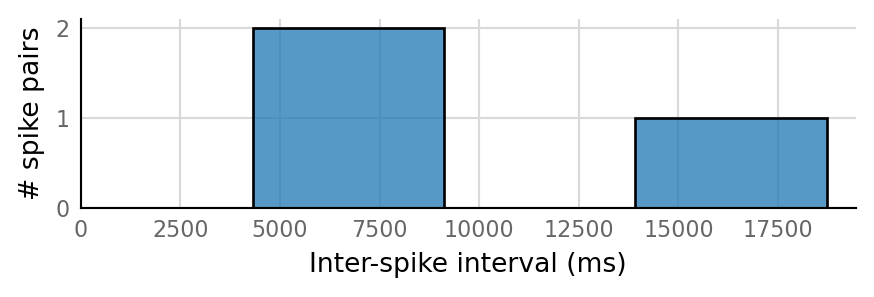
Output spike rate (1 / median ISI): 0.157 Hz
Conntest: [0.46 s]
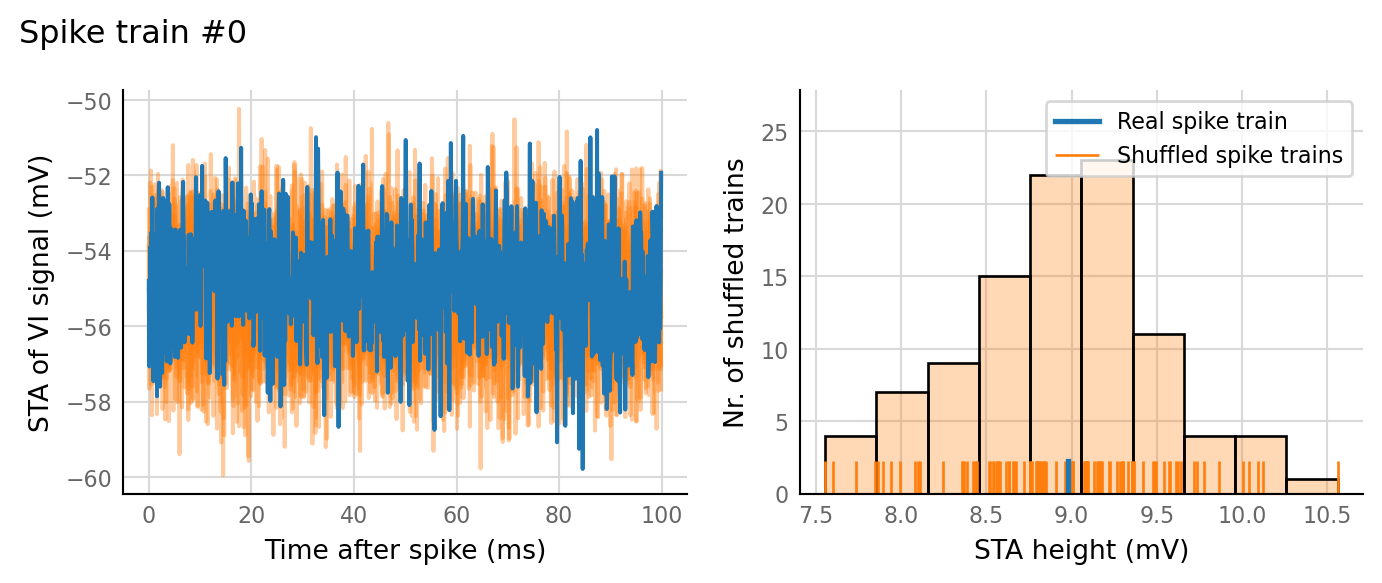
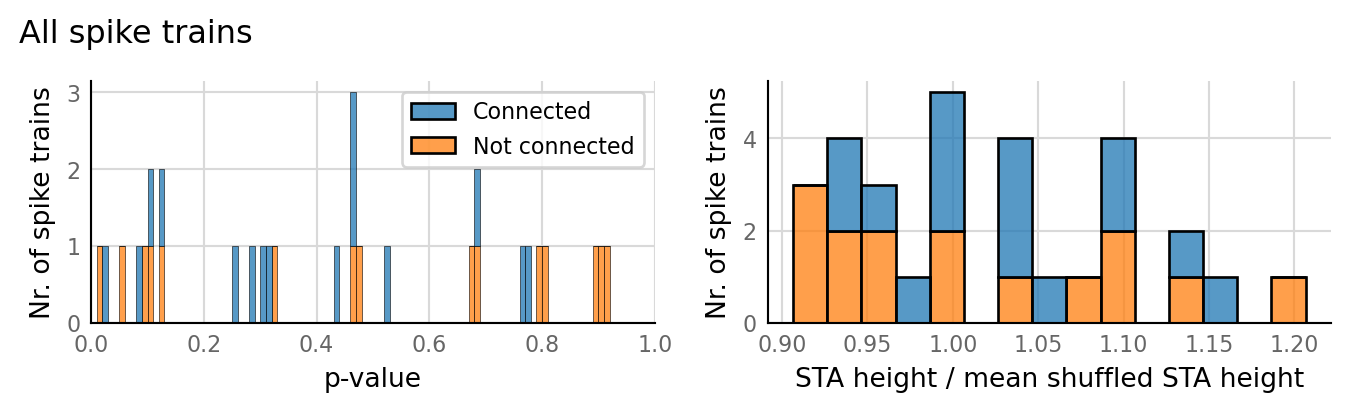
sim_eval_plot(tau_variation.paramsets[3]);
Sim: [0.05 s]
Output spike rate (1 / median ISI): 28.3 Hz
Conntest: [0.49 s]
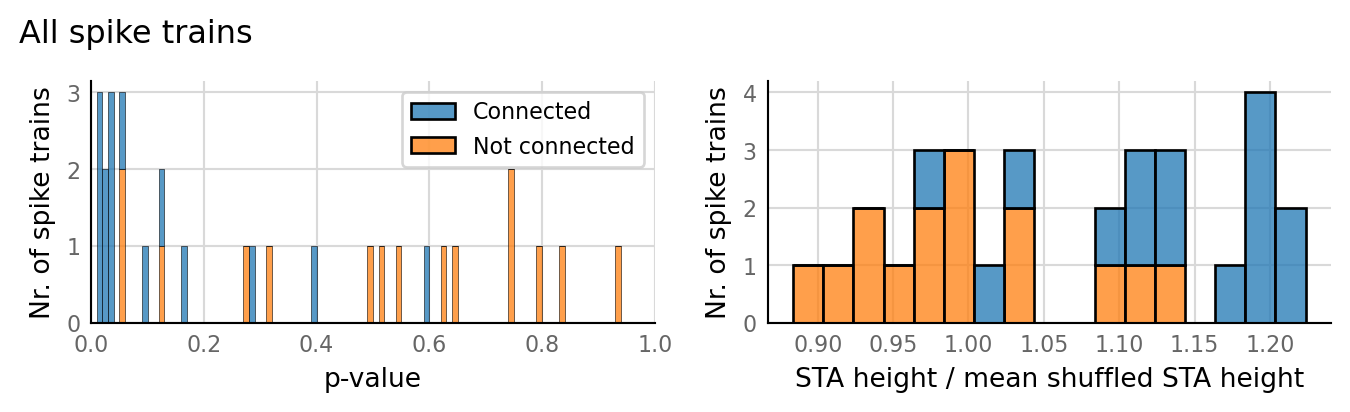
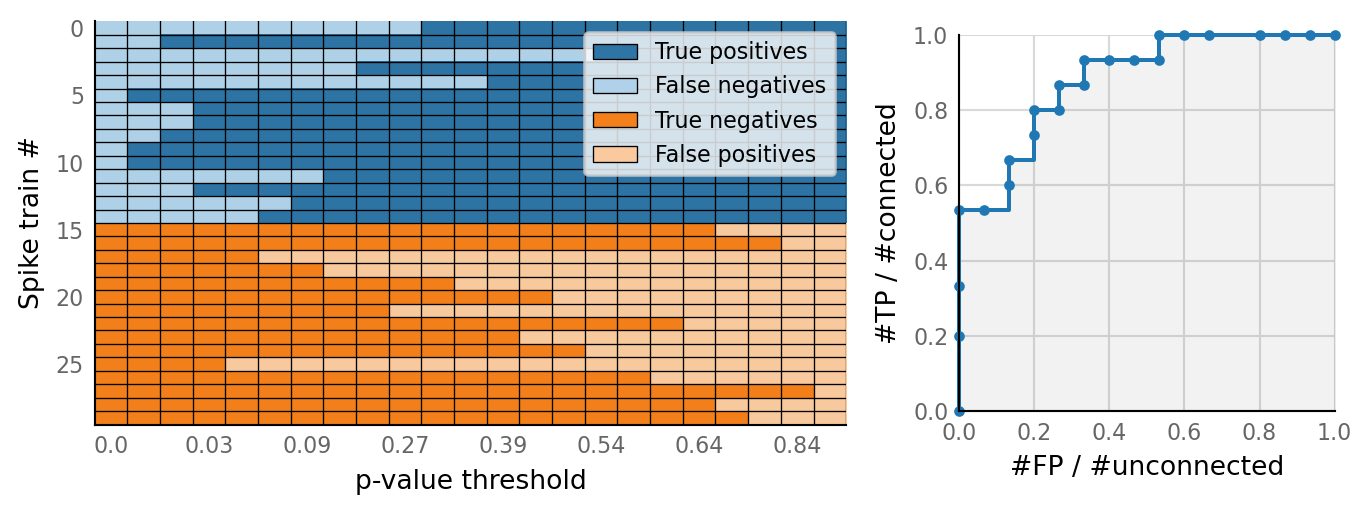
sim_eval_plot(tau_variation.paramsets[4]);
Sim: [0.04 s]
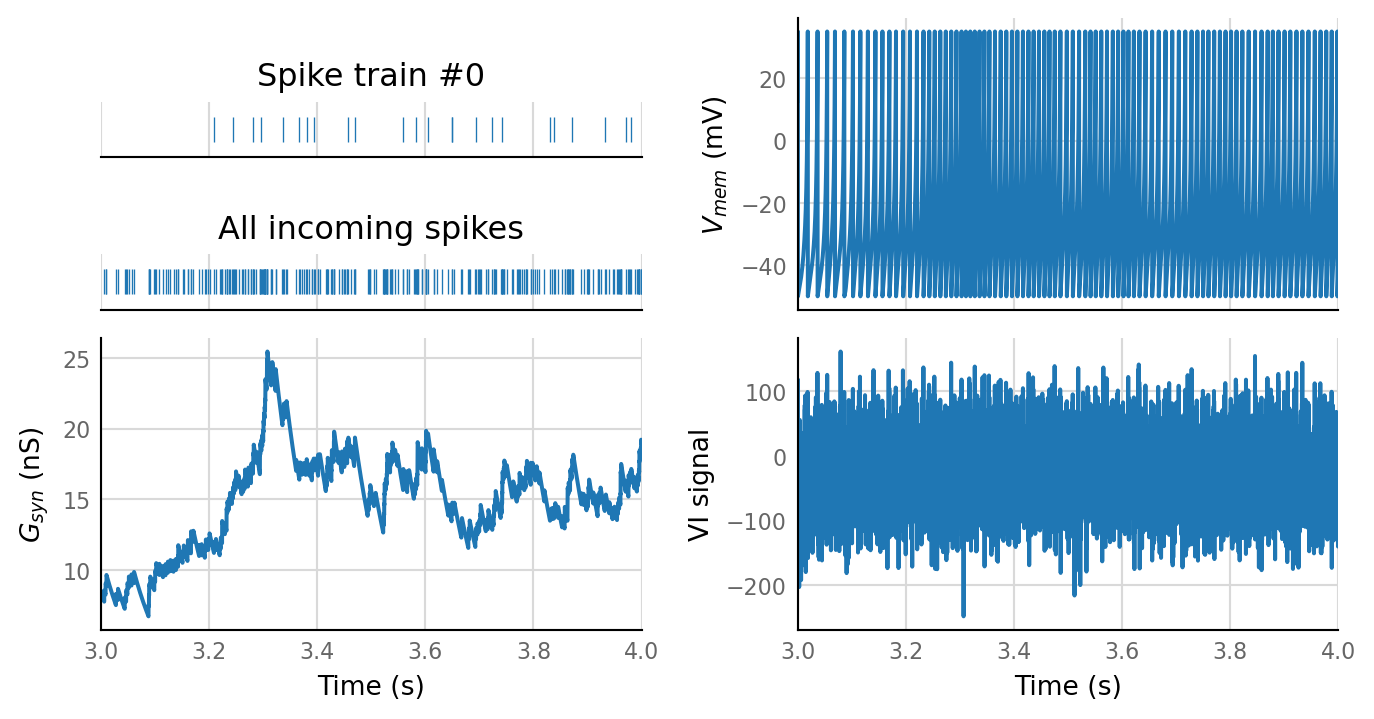
Output spike rate (1 / median ISI): 98 Hz
Conntest: [0.42 s]
Discussion¶
This again seems correlated with output spike rate.
Although: \(\tau_{syn}\) = 14 ms yields more than twice the output spike rate as the default of \(\tau_{syn}\) = 7 ms, but their detection performance is similar.
Compare with the # spike trains variation: both with 60 spike trains and with \(\tau_{syn}\) = 14 ms, the output spike rate is about 27 Hz.
However, the \(\tau_{syn}\) = 14 ms yields better detection performance (ROC-AUC of .88 – on par with baseline; vs .72).
Vary RNG seed¶
Let’s see the spread of the baseline.
seed_variation = ParamVariation(
param="rng_seed",
param_label="RNG seed",
values=(1800, 0, 1, 2, 3, 4, 5),
);
%time seed_sweeps = sim_and_eval_variations(seed_variation);
RNG seed: 1800 | Sim: [0.04 s] | Conntest: [0.05 s]
RNG seed: 0 | Sim: [0.05 s] | Conntest: [0.04 s]
RNG seed: 1 | Sim: [0.04 s] | Conntest: [0.04 s]
RNG seed: 2 | Sim: [0.06 s] | Conntest: [0.05 s]
RNG seed: 3 | Sim: [0.04 s] | Conntest: [0.04 s]
RNG seed: 4 | Sim: [0.04 s] | Conntest: [0.05 s]
RNG seed: 5 | Sim: [0.04 s] | Conntest: [0.05 s]
Wall time: 707 ms
plot_sweeps(seed_sweeps, seed_variation);
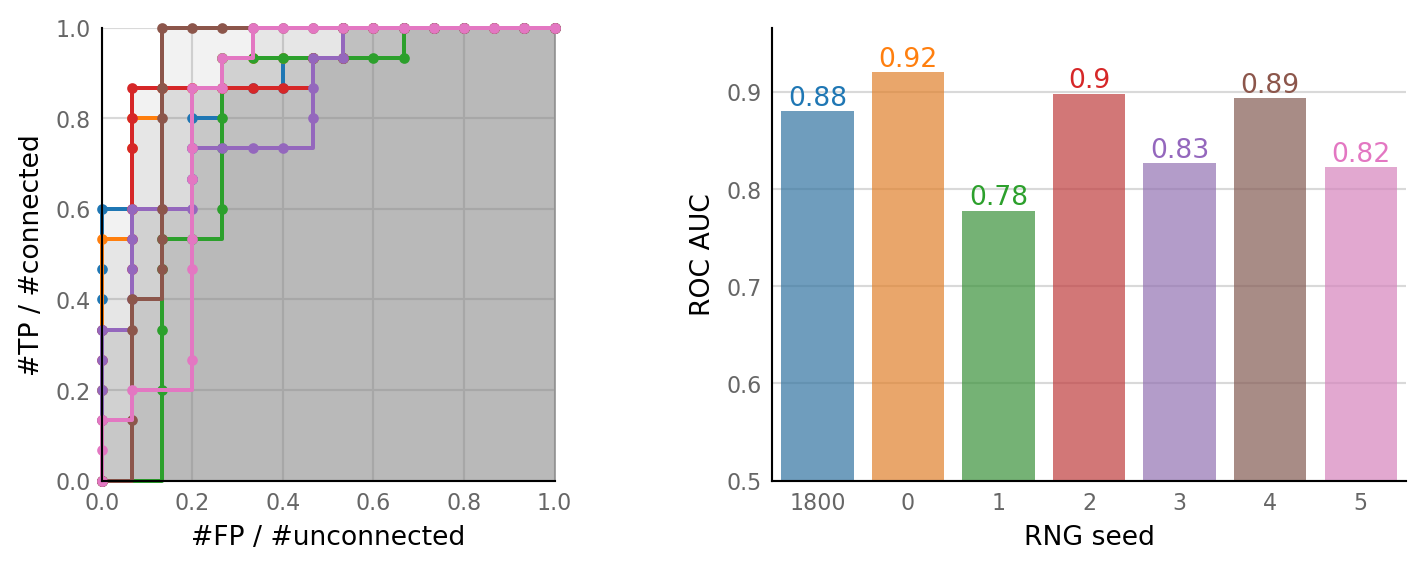
Inspect extremes¶
sim_eval_plot(seed_variation.paramsets[1]);
Sim: [0.06 s]
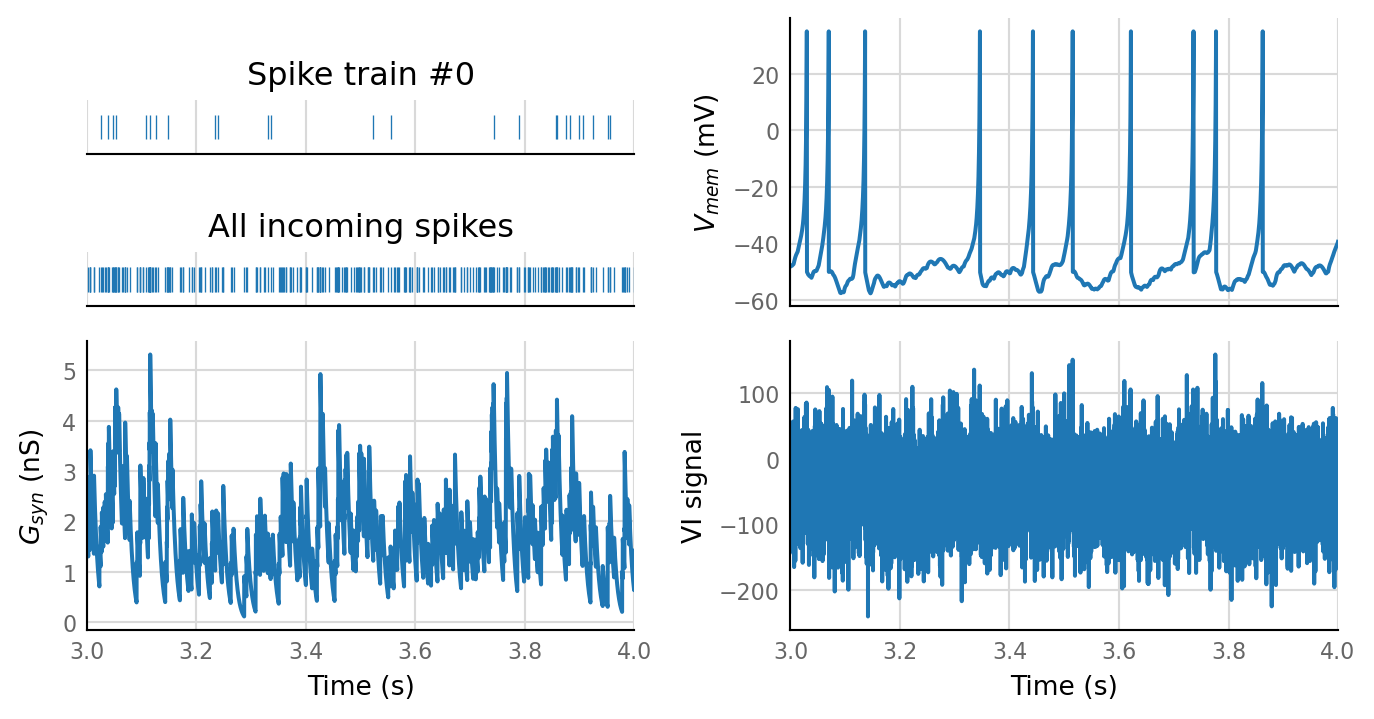
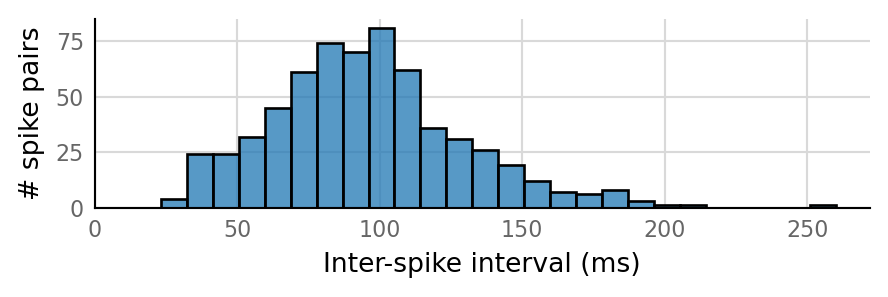
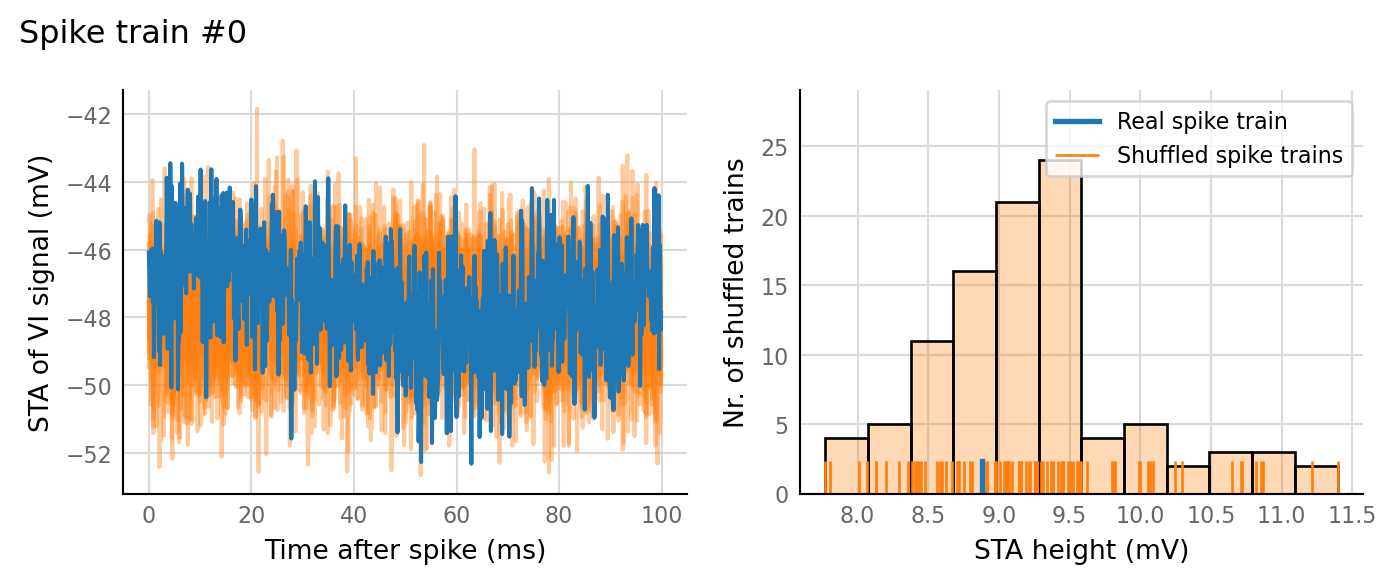
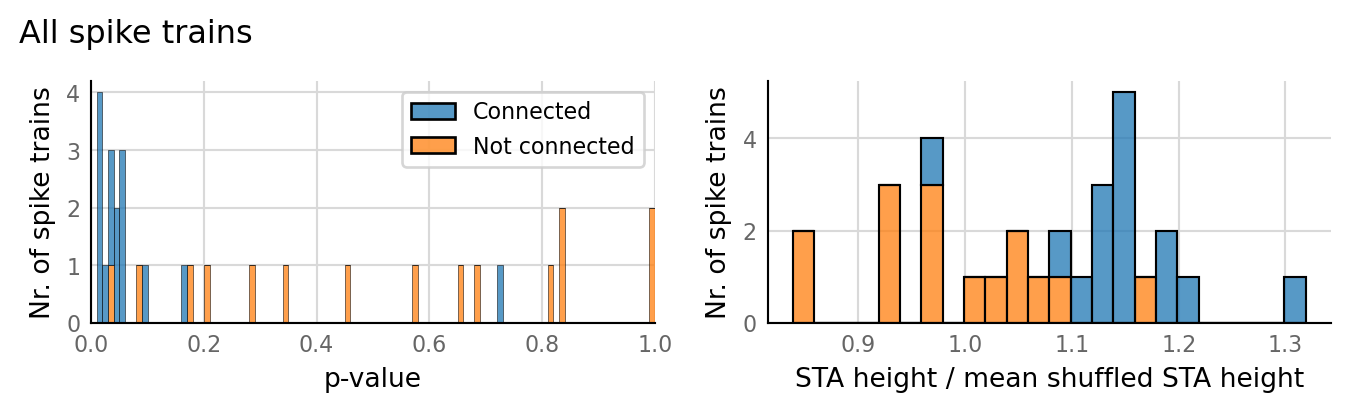
Output spike rate (1 / median ISI): 10.7 Hz
Conntest: [0.63 s]
sim_eval_plot(seed_variation.paramsets[2]);
Sim: [0.04 s]
Output spike rate (1 / median ISI): 10.9 Hz
Conntest: [0.60 s]
Reproducibility¶
v.print_reproducibility_info(verbose=True)
This cell was last run by tfiers on yoga
on Wed 27 Jan 2021, at 18:16 (UTC+0100).
Last git commit (Wed 27 Jan 2021, 18:08).
No uncommitted changes.
Platform:
Windows-10
CPython 3.8.6 (C:\conda\python.exe)
Intel(R) Core(TM) i7-10510U CPU @ 1.80GHz
Dependencies of voltage_to_wiring_sim and their installed versions:
numpy 1.19.5
matplotlib 3.3.3
numba 0.52.0
joblib 1.0.0
seaborn 0.11.1
scipy 1.6.0
preload 2.2
nptyping 1.4.0
Full conda list:
# packages in environment at C:\conda:
#
# Name Version Build Channel
anyio 2.0.2 py38haa244fe_4 conda-forge
appdirs 1.4.4 pyh9f0ad1d_0 conda-forge
argon2-cffi 20.1.0 py38h294d835_2 conda-forge
async_generator 1.10 py_0 conda-forge
atomicwrites 1.4.0 pyh9f0ad1d_0 conda-forge
attrs 20.3.0 pyhd3deb0d_0 conda-forge
babel 2.9.0 pyhd3deb0d_0 conda-forge
backcall 0.2.0 pyh9f0ad1d_0 conda-forge
backports 1.0 py_2 conda-forge
backports.functools_lru_cache 1.6.1 py_0 conda-forge
black 20.8b1 py_1 conda-forge
bleach 3.2.2 pyh44b312d_0 conda-forge
bokeh 2.2.3 py38haa244fe_0 conda-forge
brotlipy 0.7.0 py38h294d835_1001 conda-forge
bzip2 1.0.8 h8ffe710_4 conda-forge
ca-certificates 2020.12.5 h5b45459_0 conda-forge
certifi 2020.12.5 py38haa244fe_1 conda-forge
cffi 1.14.4 py38hd8c33c5_1 conda-forge
chardet 4.0.0 py38haa244fe_1 conda-forge
click 7.1.2 pyh9f0ad1d_0 conda-forge
cloudpickle 1.6.0 py_0 conda-forge
colorama 0.4.4 pyh9f0ad1d_0 conda-forge
conda 4.9.2 py38haa244fe_0 conda-forge
conda-package-handling 1.7.2 py38h8934438_0 conda-forge
console_shortcut 0.1.1 4
cryptography 3.3.1 py38hd8c33c5_1 conda-forge
cycler 0.10.0 py_2 conda-forge
cytoolz 0.11.0 py38h294d835_3 conda-forge
dask 2021.1.0 pyhd8ed1ab_0 conda-forge
dask-core 2021.1.0 pyhd8ed1ab_0 conda-forge
dataclasses 0.7 pyhb2cacf7_7 conda-forge
decorator 4.4.2 py_0 conda-forge
defusedxml 0.6.0 py_0 conda-forge
distributed 2021.1.1 py38haa244fe_0 conda-forge
docutils 0.16 pypi_0 pypi
entrypoints 0.3 pyhd8ed1ab_1003 conda-forge
freetype 2.10.4 h546665d_1 conda-forge
fsspec 0.8.5 pyhd8ed1ab_0 conda-forge
gitdb 4.0.5 py_0 conda-forge
gitpython 3.1.12 pyhd8ed1ab_0 conda-forge
heapdict 1.0.1 py_0 conda-forge
icu 68.1 h0e60522_0 conda-forge
idna 2.10 pyh9f0ad1d_0 conda-forge
importlib-metadata 3.4.0 py38haa244fe_0 conda-forge
importlib_metadata 3.4.0 hd8ed1ab_0 conda-forge
iniconfig 1.1.1 pyh9f0ad1d_0 conda-forge
intel-openmp 2020.3 h57928b3_311 conda-forge
ipykernel 5.4.2 py38h7b7c402_0 conda-forge
ipython 7.19.0 py38hc5df569_2 conda-forge
ipython_genutils 0.2.0 py_1 conda-forge
ipywidgets 7.6.3 pyhd3deb0d_0 conda-forge
jedi 0.17.2 py38haa244fe_1 conda-forge
jinja2 2.11.2 pyh9f0ad1d_0 conda-forge
joblib 1.0.0 pyhd8ed1ab_0 conda-forge
jpeg 9d h8ffe710_0 conda-forge
json5 0.9.5 pyh9f0ad1d_0 conda-forge
jsonschema 3.2.0 py_2 conda-forge
jupyter_client 6.1.11 pyhd8ed1ab_1 conda-forge
jupyter_contrib_core 0.3.3 py_2 conda-forge
jupyter_contrib_nbextensions 0.5.1 py38h32f6830_1 conda-forge
jupyter_core 4.7.0 py38haa244fe_1 conda-forge
jupyter_highlight_selected_word 0.2.0 py38haa244fe_1002 conda-forge
jupyter_latex_envs 1.4.6 py38h32f6830_1001 conda-forge
jupyter_nbextensions_configurator 0.4.1 py38haa244fe_2 conda-forge
jupyter_server 1.2.2 py38haa244fe_1 conda-forge
jupyterlab 3.0.5 pyhd8ed1ab_0 conda-forge
jupyterlab-classic 0.1.3 pyhd8ed1ab_0 conda-forge
jupyterlab-sublime 0.4.1 pypi_0 pypi
jupyterlab_pygments 0.1.2 pyh9f0ad1d_0 conda-forge
jupyterlab_server 2.1.2 pyhd8ed1ab_0 conda-forge
jupyterlab_widgets 1.0.0 pyhd8ed1ab_1 conda-forge
jupytray 0.7.post10+dirty dev_0 <develop>
keyring 21.8.0 pypi_0 pypi
kiwisolver 1.3.1 py38hbd9d945_1 conda-forge
krb5 1.17.2 hbae68bd_0 conda-forge
libarchive 3.5.1 h8686738_1 conda-forge
libblas 3.9.0 7_mkl conda-forge
libcblas 3.9.0 7_mkl conda-forge
libclang 11.0.1 default_h5c34c98_1 conda-forge
libcurl 7.71.1 h4b64cdc_8 conda-forge
libiconv 1.16 he774522_0 conda-forge
liblapack 3.9.0 7_mkl conda-forge
libpng 1.6.37 h1d00b33_2 conda-forge
libsodium 1.0.18 h8d14728_1 conda-forge
libsolv 0.7.17 h7755175_0 conda-forge
libssh2 1.9.0 hb06d900_5 conda-forge
libtiff 4.2.0 hc10be44_0 conda-forge
libxml2 2.9.10 hf5bbc77_3 conda-forge
libxslt 1.1.33 h65864e5_2 conda-forge
llvmlite 0.35.0 py38h57a6900_1 conda-forge
locket 0.2.0 py_2 conda-forge
lxml 4.6.2 py38h292cb97_1 conda-forge
lz4-c 1.9.3 h8ffe710_0 conda-forge
lzo 2.10 he774522_1000 conda-forge
m2w64-gcc-libgfortran 5.3.0 6 conda-forge
m2w64-gcc-libs 5.3.0 7 conda-forge
m2w64-gcc-libs-core 5.3.0 7 conda-forge
m2w64-gmp 6.1.0 2 conda-forge
m2w64-libwinpthread-git 5.0.0.4634.697f757 2 conda-forge
mamba 0.7.8 py38hdd88130_0 conda-forge
markupsafe 1.1.1 py38h294d835_3 conda-forge
matplotlib-base 3.3.3 py38h34ddff4_0 conda-forge
menuinst 1.4.16 py38he774522_1
mistune 0.8.4 py38h294d835_1003 conda-forge
mkl 2020.4 hb70f87d_311 conda-forge
more-itertools 8.6.0 pyhd8ed1ab_0 conda-forge
mpmath 1.1.0 py_0 conda-forge
msgpack-python 1.0.2 py38hbd9d945_1 conda-forge
msys2-conda-epoch 20160418 1 conda-forge
mypy_extensions 0.4.3 py38haa244fe_3 conda-forge
nb_conda_kernels 2.3.1 py38haa244fe_0 conda-forge
nbclassic 0.2.6 pyhd8ed1ab_0 conda-forge
nbclient 0.5.1 py_0 conda-forge
nbconvert 6.0.7 py38haa244fe_3 conda-forge
nbdime 2.1.0 py_0 conda-forge
nbformat 5.1.2 pyhd8ed1ab_1 conda-forge
nest-asyncio 1.4.3 pyhd8ed1ab_0 conda-forge
notebook 6.2.0 py38haa244fe_0 conda-forge
nptyping 1.4.0 pypi_0 pypi
numba 0.52.0 py38h4c96930_0 conda-forge
numpy 1.19.5 py38h0cc643e_1 conda-forge
olefile 0.46 pyh9f0ad1d_1 conda-forge
openssl 1.1.1i h8ffe710_0 conda-forge
packaging 20.8 pyhd3deb0d_0 conda-forge
pandas 1.2.1 py38h4c96930_0 conda-forge
pandoc 2.11.4 h8ffe710_0 conda-forge
pandocfilters 1.4.2 py_1 conda-forge
parso 0.7.1 pyh9f0ad1d_0 conda-forge
partd 1.1.0 py_0 conda-forge
pathspec 0.8.1 pyhd3deb0d_0 conda-forge
patsy 0.5.1 py_0 conda-forge
pickleshare 0.7.5 py_1003 conda-forge
pillow 8.1.0 py38hf7ce48b_1 conda-forge
pip 21.0 pyhd8ed1ab_0 conda-forge
pkginfo 1.6.1 pypi_0 pypi
pluggy 0.13.1 py38haa244fe_4 conda-forge
powershell_shortcut 0.0.1 3
preload 2.2 pypi_0 pypi
prometheus_client 0.9.0 pyhd3deb0d_0 conda-forge
prompt-toolkit 3.0.14 pyha770c72_0 conda-forge
psutil 5.8.0 py38h294d835_1 conda-forge
puprelease 1.6.1 dev_0 <develop>
py 1.10.0 pyhd3deb0d_0 conda-forge
py-cpuinfo 7.0.0 pypi_0 pypi
pycosat 0.6.3 py38h294d835_1006 conda-forge
pycparser 2.20 pyh9f0ad1d_2 conda-forge
pygments 2.7.4 pyhd8ed1ab_0 conda-forge
pyopenssl 20.0.1 pyhd8ed1ab_0 conda-forge
pyparsing 2.4.7 pyh9f0ad1d_0 conda-forge
pyqt 5.12.3 py38haa244fe_7 conda-forge
pyqt-impl 5.12.3 py38h885f38d_7 conda-forge
pyqt5-sip 4.19.18 py38h885f38d_7 conda-forge
pyqtchart 5.12 py38h885f38d_7 conda-forge
pyqtwebengine 5.12.1 py38h885f38d_7 conda-forge
pyrsistent 0.17.3 py38h294d835_2 conda-forge
pysocks 1.7.1 py38haa244fe_3 conda-forge
pytest 6.2.1 py38haa244fe_1 conda-forge
python 3.8.6 h7840368_4_cpython conda-forge
python-dateutil 2.8.1 py_0 conda-forge
python_abi 3.8 1_cp38 conda-forge
pytz 2020.5 pyhd8ed1ab_0 conda-forge
pywin32 300 py38h294d835_0 conda-forge
pywin32-ctypes 0.2.0 pypi_0 pypi
pywinpty 0.5.7 py38h32f6830_1 conda-forge
pyyaml 5.4.1 py38h294d835_0 conda-forge
pyzmq 21.0.1 py38h7a0e47e_0 conda-forge
qt 5.12.9 h5909a2a_3 conda-forge
readme-renderer 28.0 pypi_0 pypi
regex 2020.11.13 py38h294d835_1 conda-forge
reproc 14.2.1 h8ffe710_0 conda-forge
reproc-cpp 14.2.1 h0e60522_0 conda-forge
requests 2.25.1 pyhd3deb0d_0 conda-forge
requests-toolbelt 0.9.1 pypi_0 pypi
rfc3986 1.4.0 pypi_0 pypi
ruamel_yaml 0.15.80 py38h294d835_1003 conda-forge
scikit-learn 0.24.1 py38ha09990b_0 conda-forge
scipy 1.6.0 py38h5f893b4_0 conda-forge
seaborn 0.11.1 h57928b3_0 conda-forge
seaborn-base 0.11.1 pyhd8ed1ab_0 conda-forge
send2trash 1.5.0 py_0 conda-forge
setuptools 49.6.0 py38haa244fe_3 conda-forge
setuptools-scm 5.0.1 pypi_0 pypi
six 1.15.0 pyh9f0ad1d_0 conda-forge
smmap 3.0.5 pyh44b312d_0 conda-forge
snakeviz 2.1.0 pyh9f0ad1d_0 conda-forge
sniffio 1.2.0 py38haa244fe_1 conda-forge
sortedcontainers 2.3.0 pyhd8ed1ab_0 conda-forge
sqlite 3.34.0 h8ffe710_0 conda-forge
statsmodels 0.12.1 py38h347fdf6_2 conda-forge
sympy 1.7.1 py38haa244fe_1 conda-forge
tblib 1.6.0 py_0 conda-forge
terminado 0.9.2 py38haa244fe_0 conda-forge
testpath 0.4.4 py_0 conda-forge
threadpoolctl 2.1.0 pyh5ca1d4c_0 conda-forge
tk 8.6.10 h8ffe710_1 conda-forge
toml 0.10.2 pyhd8ed1ab_0 conda-forge
toolz 0.11.1 py_0 conda-forge
tornado 6.1 py38h294d835_1 conda-forge
tqdm 4.56.0 pyhd8ed1ab_0 conda-forge
traitlets 5.0.5 py_0 conda-forge
twine 3.3.0 pypi_0 pypi
typed-ast 1.4.2 py38h294d835_0 conda-forge
typing_extensions 3.7.4.3 py_0 conda-forge
typish 1.9.1 pypi_0 pypi
unitlib 0.3 pypi_0 pypi
urllib3 1.26.2 pyhd8ed1ab_0 conda-forge
vc 14.2 hb210afc_2 conda-forge
voltage-to-wiring-sim 0.1 dev_0 <develop>
vs2015_runtime 14.28.29325 h5e1d092_0 conda-forge
wcwidth 0.2.5 pyh9f0ad1d_2 conda-forge
webencodings 0.5.1 py_1 conda-forge
wheel 0.36.2 pyhd3deb0d_0 conda-forge
widgetsnbextension 3.5.1 py38haa244fe_4 conda-forge
win_inet_pton 1.1.0 py38haa244fe_2 conda-forge
wincertstore 0.2 py38haa244fe_1006 conda-forge
windows-curses 2.2.0 pypi_0 pypi
winpty 0.4.3 4 conda-forge
winshell 0.6 pypi_0 pypi
xz 5.2.5 h62dcd97_1 conda-forge
yaml 0.2.5 he774522_0 conda-forge
zeromq 4.3.3 h0e60522_3 conda-forge
zict 2.0.0 py_0 conda-forge
zipp 3.4.0 py_0 conda-forge
zlib 1.2.11 h62dcd97_1010 conda-forge
zstd 1.4.8 h4e2f164_1 conda-forge