2021-12-06 • Local shape of \dot{V} in 2D HH
Contents
2021-12-06 • Local shape of \(\dot{V}\) in 2D HH¶
What’s the shape of dV/dt (V) for fixed \(n\) in the Hodgkin-Huxley model simplified to 2D (as done in Izhikevich book sec 5.2 or here in the epfl book)?
[Prelude]¶
from voltage_to_wiring_sim.notebook_init import *
Preloading: numpy, numba, matplotlib.pyplot, seaborn.
Importing from submodules … ✔
Imported `np`, `mpl`, `plt`, `sns`, `pd`
Imported codebase (`voltage_to_wiring_sim`) as `v`
Imported `*` from `v.support.units`
Setup autoreload
v.print_reproducibility_info()
This cell was last run by lpxtf3 on DUIP74576 on Wed 08 Dec 2021, at 19:15 (UTC+0000).
Last git commit (Tue 07 Dec 2021, 03:59).
Uncommited changes to 4 files.
Params¶
From HH1952 / Izh book sec 2.3.1.
(Alternatively, params could be taken from Ch 2.2 of the epfl book, which has HH params fitted to a cortical pyramidal neuron, instead of params for the squid giant axon used here).
E_shift = 65 * mV;
c = 1 * (μF / cm**2)
E_K = -12 * mV - E_shift
E_Na = 120 * mV - E_shift
E_L = 10.6 * mV - E_shift
g_K = 36 * (mS / cm**2)
g_Na = 120 * (mS / cm**2)
g_L = 0.03 * (mS / cm**2)
α_m = lambda V: 0.1 * (25 - V/mV) / (np.exp((25-V/mV)/10) - 1) / ms
β_m = lambda V: 4 * np.exp(-V/mV/18) / ms
m_inf = lambda V: α_m(V) / (α_m(V) + β_m(V))
α_n = lambda V: 0.01 * (10 - V/mV) / (np.exp((10-V/mV)/10) - 1) / ms
β_n = lambda V: 0.125 * np.exp(-V/mV/80) / ms
n_inf = lambda V: α_n(V) / (α_n(V) + β_n(V))
τ_n = lambda V: 1 / (α_n(V) + β_n(V));
Simplified Hodgkin-Huxley model¶
With \(τ_m = 0\) and \(n(t) \propto h(t)\), the 4D system is reduced to just 2D.
h = lambda n: 0.89 - 1.1 * n
I_K = lambda V, n: - g_K * n**4 * (V - E_K)
I_Na = lambda V, n: - g_Na * m_inf(V)**3 * h(n) * (V - E_Na)
I_L = lambda V: - g_L * (V - E_L)
I_tot = lambda V, n, I: I + I_K(V,n) + I_Na(V,n) + I_L(V)
dV_dt = lambda V, n, I: I_tot(V,n,I) / c
dn_dt = lambda V, n: (n_inf(V) - n) / τ_n(V);
The equation for \(\dot{n}\) is only used in the simulation at the end. The other sections all analyse \(\dot{V}\) independently from \(\dot{n}\).
\(dV/dt\)¶
def horizontal_ylabel(ax, text, dx=-6, dy=-2, ha="right", x=0, y=1, bg_pad=0.4, **text_kwargs):
""" dx and dy are in points, i.e. 1/72th of an inch. """
offset = mpl.transforms.ScaledTranslation(dx/72, dy/72, ax.figure.dpi_scale_trans)
bbox=dict(facecolor=ax.get_facecolor(), edgecolor='none', boxstyle=f'square,pad={bg_pad}')
return ax.text(x, y, text, transform=ax.transAxes + offset,
ha=ha, va="top", bbox=bbox, **text_kwargs)
def make_V_dV_ax(V_min=-100*mV, V_max=5*mV):
fig, ax = plt.subplots()
ax.axhline(y=0, color='k')
ax.set_xlabel("$V$ (mV)")
horizontal_ylabel(ax, "$\dot{V}$ (mV/mS)")
V = np.linspace(V_min, V_max, num=1000)
return ax, V
ax, V = make_V_dV_ax()
ax.plot(V / mV, dV_dt(V, n=0.0, I=0) / (mV / ms));
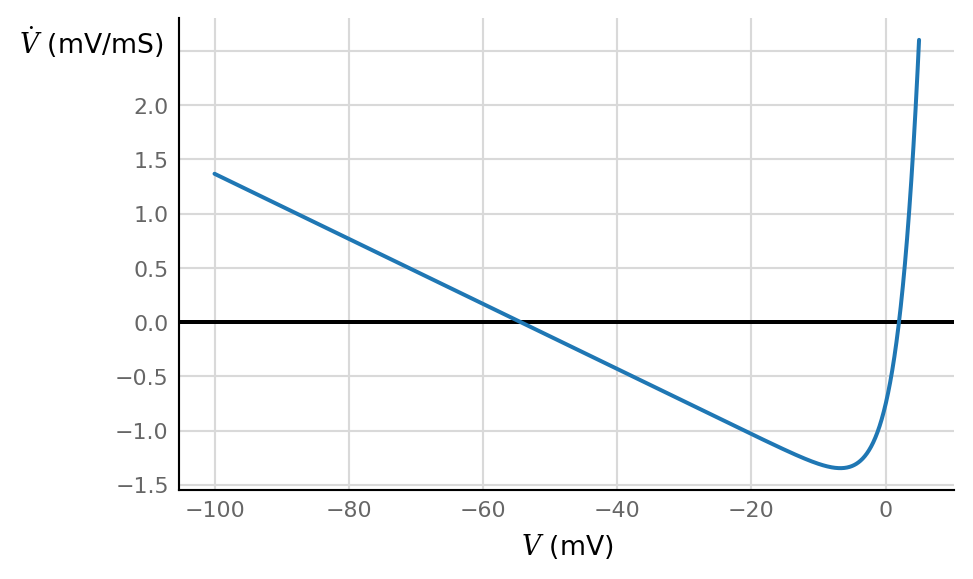
from scipy.optimize import newton
f = partial(dV_dt, n=0, I=0)
newton(f, [-60*mV, 0*mV]) / mV
array([-54.4, 2.007])
There is a stable node at \(-\)54.4 mV ( → ⚫ ←, resting potential)
and an unstable node at 2 mV ( ← ⚪ →, spike threshold).
This \(\dot{V}\) function has the same shape as in the AdEx (aka aEIF) model: linear + exponential:
(This is eq 6.3 here, where we equated the membrane conductance \(1/R\) with Izhikevich’s \(k\)).
It is not the same shape as in the Izhikevich model. There, \(\dot{V}\) is quadratic:
(\(w\) is Izhikevich’s \(u\), is the 2D HH model’s \(n\)).
Izhikevich’s \(dV/dt\)¶
C = 1320 * pF # 100 pF
k = 0.7 * (nS/mV)
V_r = -54.4 * mV # -65 mV
V_t = 2 * mV # -40 mV
dV_dt_Izh = lambda V: (k * (V - V_r) * (V - V_t)) / C
ax, V = make_V_dV_ax(V_max=20*mV)
# ax, V = make_V_dV_ax(V_min=-54.5*mV, V_max=-54.3*mV)
ax.plot(V / mV, dV_dt_Izh(V) / (mV / ms), label="Izhikevich")
ax.plot(V / mV, dV_dt(V,0,0) / (mV / ms), label="HH 2D")
ax.set_xlim(-100, 20)
ax.set_ylim(-1.7, 4.9)
label_lines(ax.lines, xvals=[-88, -18], outline_width=7);
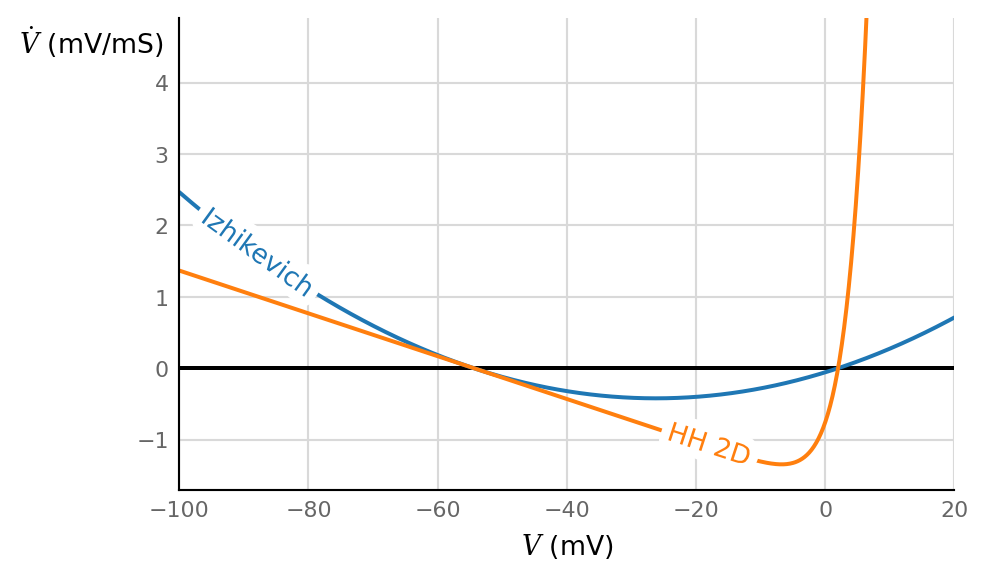
The parabola’s zeros (\(V_r\) and \(V_t\)) were chosen to match those of the 2D Hodgkin-Huxley model.
The capacitance \(C\) was chosen so that the slopes of the two models approximately match at the resting potential.
Input current \(I\)¶
ax, V = make_V_dV_ax(V_max = 10*mV)
for I in np.array([0, -10, 10, 20]) * nA/mm**2:
ax.plot(V / mV, dV_dt(V, n=0.0, I=I) / (mV / ms), label=f"$I = {I/(nA/mm**2):g}$ nA/mm²")
ax.set_xlim(-83, 10)
ax.set_ylim(-2.8, 3.1)
label_lines(ax.lines, xvals=[-38]*4, outline_width=7);
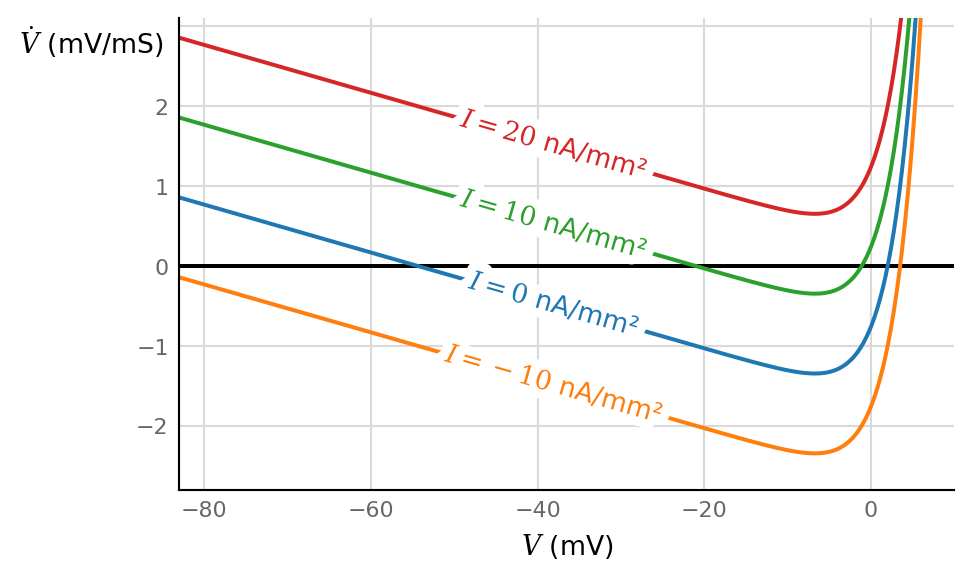
Negative input current → lower stable node aka resting potential.
Somewhere between +10 and +20 nA/mm², the stable node and the unstable node (‘saddle’) join and disappear (‘annihilate’); this is the saddle–node AKA fold bifurcation. There are no more fixed points, the neuron spikes continuously. (Thanks to a voltage return/reset not visible here).
Slow current gating variable \(n\)¶
ax, V = make_V_dV_ax(V_max = 10*mV)
for n in [0, 0.1, 0.2, 0.3]:
ax.plot(V / mV, dV_dt(V, n, I=0) / (mV / ms), label=f"$n = {n}$")
ax.set_xlim(-100, 10)
labels = label_lines(ax.lines); # labels after lims, so rotations are correct (issue 36)
labels[0].set_position((-50, 0))
labels[1].set_position((-32, -1.1))
labels[2].set_position((-23, -4))
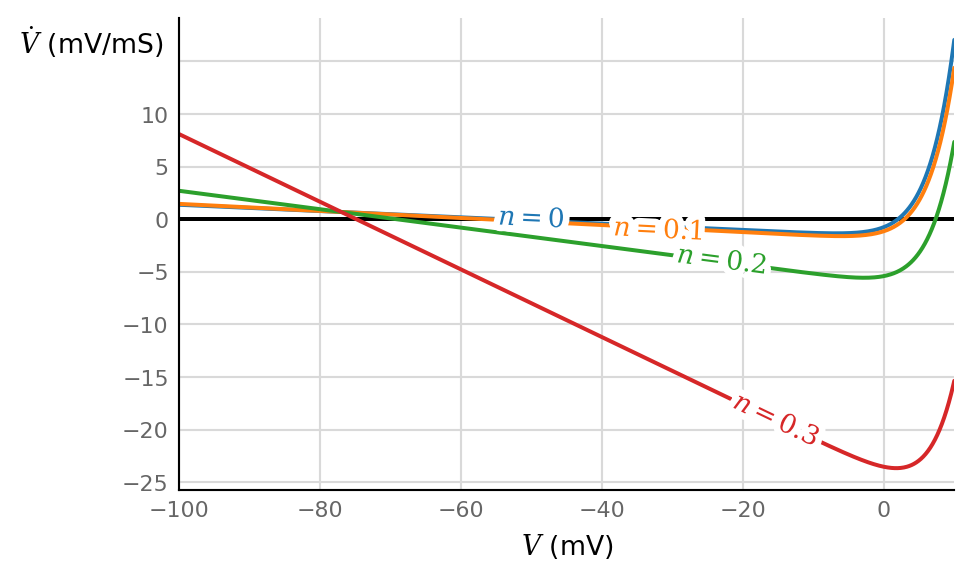
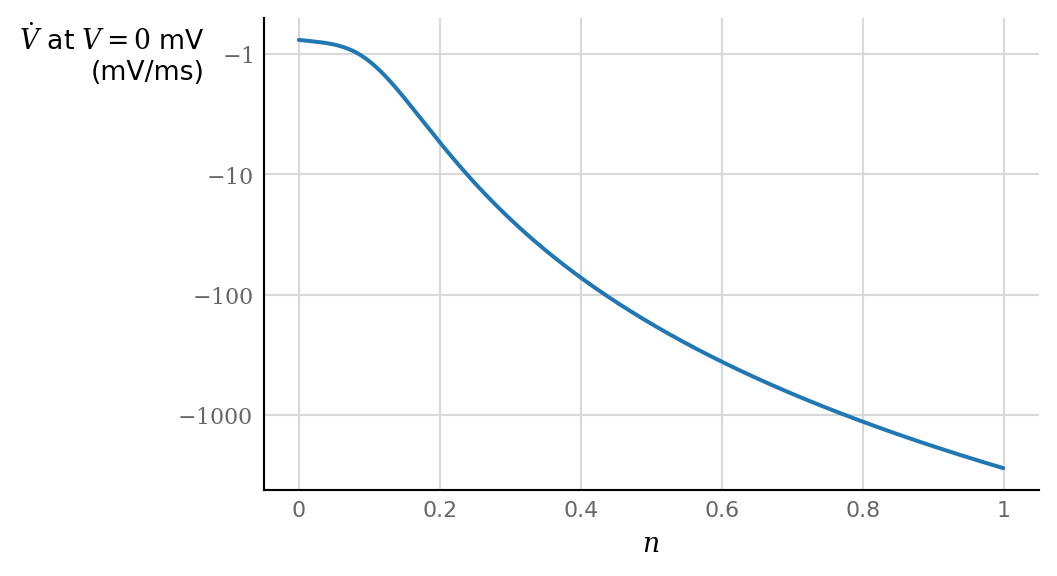
\(n ∈ (0, 1)\) (it is a gating variable).
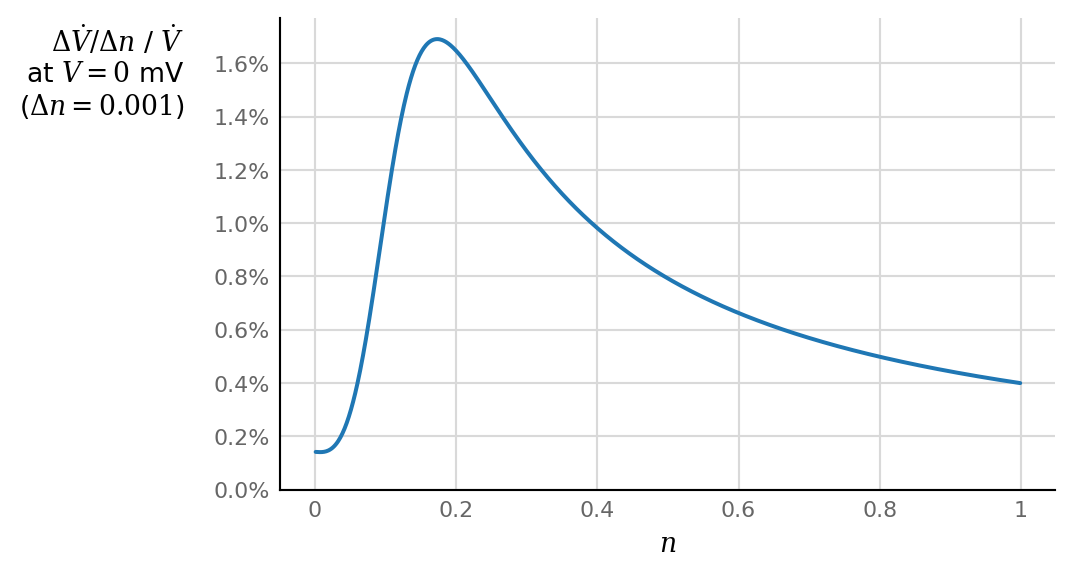
Note the weak influence of lower values of \(n\), but the strongly growing influence for higher values, from \(n > ± 0.1\) onwards.
dn = 0.001
n = np.linspace(0, 1, num=round(1/dn), endpoint=False)
fig, ax = plt.subplots()
Vdot = dV_dt(0 * mV, n, I=0) / (mV/ms)
ax.plot(n, Vdot)
# ax.set_xscale("log")
ax.set_yscale("symlog", linthresh=1e-6) # this is just -(log(-x))
ax.set_xlabel("$n$")
horizontal_ylabel(ax, "$\dot{V}$ at $V = 0$ mV"+"\n"+f"(mV/ms)", dx=-22)
ax.xaxis.set_major_formatter(mpl.ticker.FormatStrFormatter("%g"));
ax.yaxis.set_major_formatter(mpl.ticker.FormatStrFormatter("$%g$"));
fig, ax = plt.subplots()
l = ax.plot(n[1:], np.diff(Vdot) / Vdot[1:])
ax.set_xlabel("$n$")
# ax.set_xscale("log")
horizontal_ylabel(ax, "$Δ\dot{V}/Δn$ / $\dot{V}$"+"\n"+"at $V = 0$ mV"+"\n"+f"($Δn = {dn:g}$)", dx=-36)
ax.set_ylim(bottom=0)
ax.yaxis.set_major_formatter(mpl.ticker.PercentFormatter(xmax=1, decimals=1))
ax.xaxis.set_major_formatter(mpl.ticker.FormatStrFormatter("%g"));
Simulation¶
Time
duration = 2 * second
# duration = 200 * ms
Δt = 0.01 * ms
N = round(duration / Δt)
t = np.linspace(0, duration / ms, N, endpoint=False);
Input
from scipy.signal.windows import gaussian
np.random.seed(1)
I = np.random.randn(N) * 1400 * nA/mm**2
σ = 6 * ms
M = round(2 * 4 * σ / Δt)
I = np.convolve(I, gaussian(M, std=σ / Δt) / M, "same"); # local weighted average, to smooth
# I = np.ones(N) * 60 * nA/mm**2;
ODE integration
V = np.ones(N) * -54.4 * mV
n = n_inf(V)
for i in range(N - 1):
V[i+1] = V[i] + dV_dt(V[i], n[i], I[i]) * Δt
n[i+1] = n[i] + dn_dt(V[i], n[i]) * Δt
nrows = 4
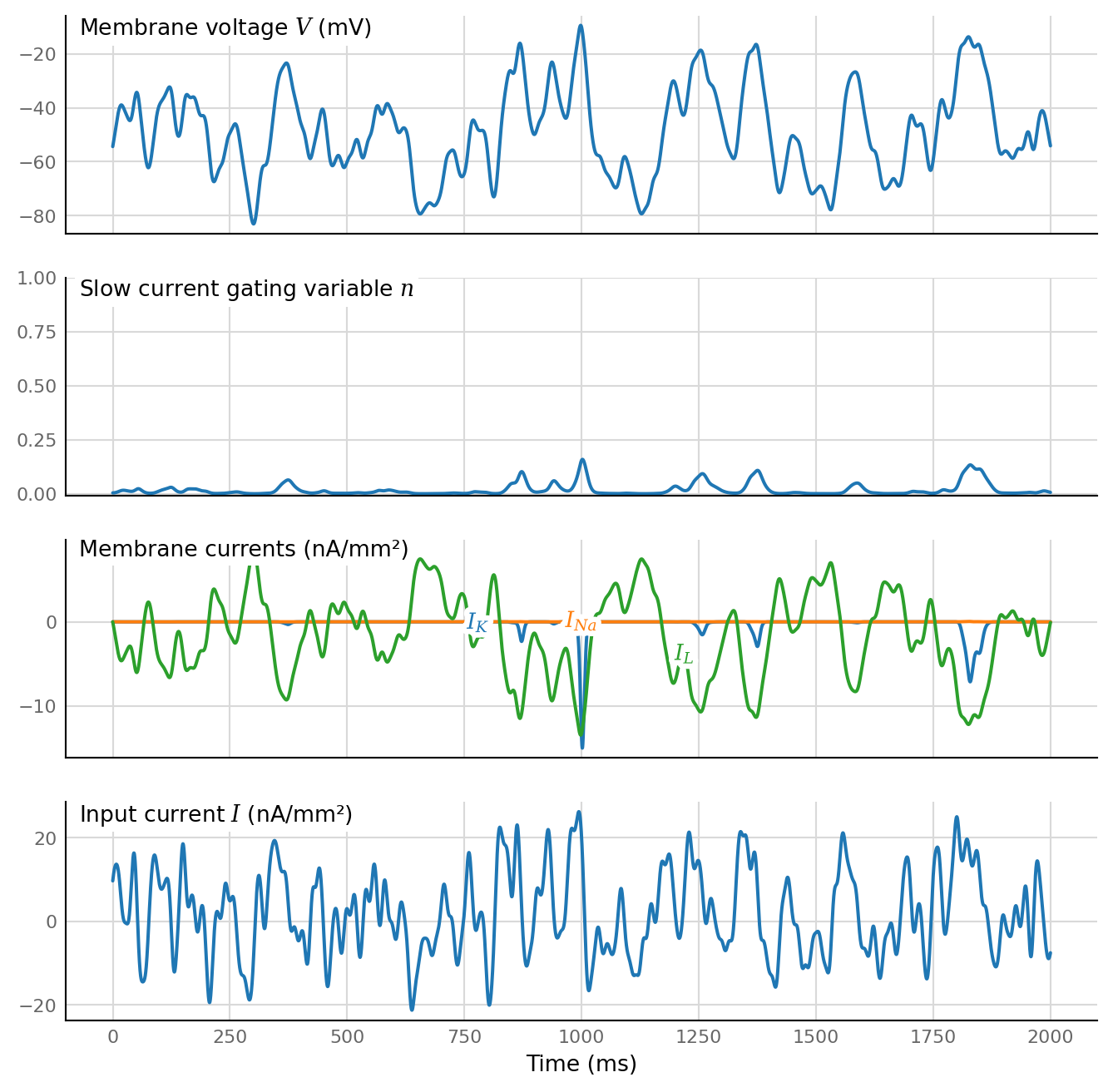
fig, axs = plt.subplots(nrows, **v.figsize(width=800, aspect=1), sharex=True)
ylabel_inside = partial(horizontal_ylabel, ha="left", dx=6, dy=0, bg_pad=0.2)
axs[0].plot(t, V / mV)
ylabel_inside(axs[0], "Membrane voltage $V$ (mV)")
axs[1].plot(t, n)
axs[1].set_ylim(-0.01, 1)
ylabel_inside(axs[1], "Slow current gating variable $n$")
axs[2].plot(t, I_K(V, n) / (nA/mm**2), label="$I_{K}$")
axs[2].plot(t, I_Na(V, n) / (nA/mm**2), label="$I_{Na}$")
axs[2].plot(t, I_L(V) / (nA/mm**2), label="$I_{L}$")
# axs[2].set_ylim(-32, 32)
ylabel_inside(axs[2], "Membrane currents (nA/mm²)")
label_lines(axs[2].lines, align=False, shrink_factor=0.3)
# label_lines(axs[2].lines, xvals=[620, 60, 140], align=False)
axs[3].plot(t, I / (nA/mm**2))
ylabel_inside(axs[3], "Input current $I$ (nA/mm²)")
axs[3].set_xlabel("Time (ms)");
[Reproducibility]¶
v.print_reproducibility_info(verbose=True)
This cell was last run by lpxtf3 on DUIP74576
on Tue 07 Dec 2021, at 02:33 (UTC+0000).
Last git commit (Mon 06 Dec 2021, 20:02).
Uncommited changes to:
M notebooks/2021-12-06__local_HH_dV_shape.ipynb
?? notebooks/2021-11-18__constant_input_spikes.ipynb
?? notebooks/2021-11-18__prototype_new_sim.ipynb
Platform:
Windows-10
CPython 3.9.6 (C:\miniforge3\python.exe)
Intel(R) Xeon(R) W-2123 CPU @ 3.60GHz
Dependencies of voltage_to_wiring_sim and their installed versions:
numpy 1.21.1
matplotlib 3.4.2
numba 0.53.1
joblib 1.0.1
seaborn 0.11.1
scipy 1.7.0
preload 2.2
nptyping 1.4.2
Full conda list:
# packages in environment at C:\miniforge3:
#
# Name Version Build Channel
argon2-cffi 20.1.0 py39hb82d6ee_2 conda-forge
async_generator 1.10 py_0 conda-forge
attrs 21.2.0 pyhd8ed1ab_0 conda-forge
backcall 0.2.0 pyh9f0ad1d_0 conda-forge
backports 1.0 py_2 conda-forge
backports.functools_lru_cache 1.6.4 pyhd8ed1ab_0 conda-forge
black 21.9b0 pyhd8ed1ab_1 conda-forge
bleach 3.3.1 pyhd8ed1ab_0 conda-forge
brian2 2.5.0.1 py39h2e07f2f_0 conda-forge
brotlipy 0.7.0 py39hb82d6ee_1001 conda-forge
ca-certificates 2021.10.8 h5b45459_0 conda-forge
certifi 2021.10.8 py39hcbf5309_1 conda-forge
cffi 1.14.6 py39h0878f49_0 conda-forge
chardet 4.0.0 py39hcbf5309_1 conda-forge
charset-normalizer 2.0.0 pyhd8ed1ab_0 conda-forge
click 7.1.2 pypi_0 pypi
colorama 0.4.4 pyh9f0ad1d_0 conda-forge
colorful 0.5.4 pypi_0 pypi
conda 4.11.0 py39hcbf5309_0 conda-forge
conda-package-handling 1.7.3 py39hb3671d1_0 conda-forge
cryptography 3.4.7 py39hd8d06c1_0 conda-forge
cycler 0.10.0 pypi_0 pypi
cython 0.29.24 py39h415ef7b_1 conda-forge
dataclasses 0.8 pyhc8e2a94_3 conda-forge
debugpy 1.4.1 py39h415ef7b_0 conda-forge
decorator 5.0.9 pyhd8ed1ab_0 conda-forge
defusedxml 0.7.1 pyhd8ed1ab_0 conda-forge
entrypoints 0.3 pyhd8ed1ab_1003 conda-forge
gsl 2.7 hdfb1a43_0 conda-forge
icu 68.2 h0e60522_0 conda-forge
idna 3.1 pyhd3deb0d_0 conda-forge
importlib-metadata 4.6.1 py39hcbf5309_0 conda-forge
intel-openmp 2021.4.0 h57928b3_3556 conda-forge
ipykernel 6.0.3 py39h832f523_0 conda-forge
ipython 7.25.0 py39h832f523_1 conda-forge
ipython_genutils 0.2.0 py_1 conda-forge
ipywidgets 7.6.5 pyhd8ed1ab_0 conda-forge
jedi 0.18.0 py39hcbf5309_2 conda-forge
jinja2 3.0.1 pyhd8ed1ab_0 conda-forge
joblib 1.0.1 pypi_0 pypi
jpeg 9d h8ffe710_0 conda-forge
jsonschema 3.2.0 pyhd8ed1ab_3 conda-forge
jupyter_client 6.1.12 pyhd8ed1ab_0 conda-forge
jupyter_contrib_core 0.3.3 py_2 conda-forge
jupyter_contrib_nbextensions 0.5.1 pyhd8ed1ab_2 conda-forge
jupyter_core 4.7.1 py39hcbf5309_0 conda-forge
jupyter_highlight_selected_word 0.2.0 py39hcbf5309_1002 conda-forge
jupyter_latex_envs 1.4.6 pyhd8ed1ab_1002 conda-forge
jupyter_nbextensions_configurator 0.4.1 py39hcbf5309_2 conda-forge
jupyterlab_pygments 0.1.2 pyh9f0ad1d_0 conda-forge
jupyterlab_widgets 1.0.2 pyhd8ed1ab_0 conda-forge
kiwisolver 1.3.1 pypi_0 pypi
libblas 3.9.0 12_win64_mkl conda-forge
libcblas 3.9.0 12_win64_mkl conda-forge
libclang 11.1.0 default_h5c34c98_1 conda-forge
libiconv 1.16 he774522_0 conda-forge
liblapack 3.9.0 12_win64_mkl conda-forge
libpng 1.6.37 h1d00b33_2 conda-forge
libsodium 1.0.18 h8d14728_1 conda-forge
libxml2 2.9.12 hf5bbc77_0 conda-forge
libxslt 1.1.33 h65864e5_2 conda-forge
libzlib 1.2.11 h8ffe710_1013 conda-forge
llvmlite 0.36.0 pypi_0 pypi
lxml 4.6.3 py39h4fd7cdf_0 conda-forge
m2w64-gcc-libgfortran 5.3.0 6 conda-forge
m2w64-gcc-libs 5.3.0 7 conda-forge
m2w64-gcc-libs-core 5.3.0 7 conda-forge
m2w64-gmp 6.1.0 2 conda-forge
m2w64-libwinpthread-git 5.0.0.4634.697f757 2 conda-forge
markupsafe 2.0.1 py39hb82d6ee_0 conda-forge
matplotlib 3.4.2 pypi_0 pypi
matplotlib-inline 0.1.2 pyhd8ed1ab_2 conda-forge
matplotlib-label-lines 0.4.3 pypi_0 pypi
menuinst 1.4.17 py39hcbf5309_1 conda-forge
miniforge_console_shortcut 2.0 h57928b3_0 conda-forge
mistune 0.8.4 py39hb82d6ee_1004 conda-forge
mkl 2021.4.0 h0e2418a_729 conda-forge
more-itertools 8.12.0 pypi_0 pypi
mpmath 1.2.1 pyhd8ed1ab_0 conda-forge
msys2-conda-epoch 20160418 1 conda-forge
mypy_extensions 0.4.3 py39hcbf5309_4 conda-forge
nbclient 0.5.3 pyhd8ed1ab_0 conda-forge
nbconvert 6.1.0 py39hcbf5309_0 conda-forge
nbformat 5.1.3 pyhd8ed1ab_0 conda-forge
nest-asyncio 1.5.1 pyhd8ed1ab_0 conda-forge
notebook 6.4.0 pyha770c72_0 conda-forge
nptyping 1.4.2 pypi_0 pypi
numba 0.53.1 pypi_0 pypi
numpy 1.21.1 pypi_0 pypi
openssl 1.1.1l h8ffe710_0 conda-forge
packaging 21.0 pyhd8ed1ab_0 conda-forge
pandas 1.3.1 pypi_0 pypi
pandoc 2.14.1 h8ffe710_0 conda-forge
pandocfilters 1.4.2 py_1 conda-forge
parso 0.8.2 pyhd8ed1ab_0 conda-forge
pathspec 0.9.0 pyhd8ed1ab_0 conda-forge
pickleshare 0.7.5 py_1003 conda-forge
pillow 8.3.1 pypi_0 pypi
pip 21.2.1 pyhd8ed1ab_0 conda-forge
platformdirs 2.3.0 pyhd8ed1ab_0 conda-forge
preload 2.2 pypi_0 pypi
prettyprinter 0.18.0 pypi_0 pypi
prometheus_client 0.11.0 pyhd8ed1ab_0 conda-forge
prompt-toolkit 3.0.19 pyha770c72_0 conda-forge
py-cpuinfo 8.0.0 pyhd8ed1ab_0 conda-forge
pycosat 0.6.3 py39hb82d6ee_1006 conda-forge
pycparser 2.20 pyh9f0ad1d_2 conda-forge
pygments 2.9.0 pyhd8ed1ab_0 conda-forge
pympler 0.9 pypi_0 pypi
pyopenssl 20.0.1 pyhd8ed1ab_0 conda-forge
pyparsing 2.4.7 pyh9f0ad1d_0 conda-forge
pyqt 5.12.3 py39hcbf5309_7 conda-forge
pyqt-impl 5.12.3 py39h415ef7b_7 conda-forge
pyqt5-sip 4.19.18 py39h415ef7b_7 conda-forge
pyqtchart 5.12 py39h415ef7b_7 conda-forge
pyqtwebengine 5.12.1 py39h415ef7b_7 conda-forge
pyrsistent 0.17.3 py39hb82d6ee_2 conda-forge
pysocks 1.7.1 py39hcbf5309_3 conda-forge
python 3.9.6 h7840368_1_cpython conda-forge
python-dateutil 2.8.2 pyhd8ed1ab_0 conda-forge
python_abi 3.9 2_cp39 conda-forge
pytz 2021.1 pypi_0 pypi
pywin32 300 py39hb82d6ee_0 conda-forge
pywinpty 1.1.3 py39h99910a6_0 conda-forge
pyyaml 6.0 py39hb82d6ee_0 conda-forge
pyzmq 22.1.0 py39he46f08e_0 conda-forge
qt 5.12.9 h5909a2a_4 conda-forge
regex 2021.10.23 py39hb82d6ee_1 conda-forge
requests 2.26.0 pyhd8ed1ab_0 conda-forge
ruamel_yaml 0.15.80 py39hb82d6ee_1004 conda-forge
scipy 1.7.0 pypi_0 pypi
seaborn 0.11.1 pypi_0 pypi
send2trash 1.7.1 pyhd8ed1ab_0 conda-forge
setuptools 49.6.0 py39hcbf5309_3 conda-forge
six 1.16.0 pyh6c4a22f_0 conda-forge
sqlite 3.36.0 h8ffe710_0 conda-forge
sympy 1.9 py39hcbf5309_1 conda-forge
tbb 2021.4.0 h2d74725_1 conda-forge
terminado 0.10.1 py39hcbf5309_0 conda-forge
testpath 0.5.0 pyhd8ed1ab_0 conda-forge
tomli 1.2.2 pyhd8ed1ab_0 conda-forge
tornado 6.1 py39hb82d6ee_1 conda-forge
tqdm 4.61.2 pyhd8ed1ab_1 conda-forge
traitlets 5.0.5 py_0 conda-forge
typed-ast 1.4.3 py39hb82d6ee_1 conda-forge
typing_extensions 3.10.0.2 pyha770c72_0 conda-forge
typish 1.9.2 pypi_0 pypi
tzdata 2021a he74cb21_1 conda-forge
ucrt 10.0.20348.0 h57928b3_0 conda-forge
urllib3 1.26.6 pyhd8ed1ab_0 conda-forge
vc 14.2 hb210afc_5 conda-forge
voltage-to-wiring-sim 0.1 dev_0 <develop>
vs2015_runtime 14.29.30037 h902a5da_5 conda-forge
wcwidth 0.2.5 pyh9f0ad1d_2 conda-forge
webencodings 0.5.1 py_1 conda-forge
wheel 0.36.2 pyhd3deb0d_0 conda-forge
widgetsnbextension 3.5.2 py39hcbf5309_1 conda-forge
win_inet_pton 1.1.0 py39hcbf5309_2 conda-forge
wincertstore 0.2 py39hcbf5309_1006 conda-forge
winpty 0.4.3 4 conda-forge
winshell 0.6 pypi_0 pypi
yaml 0.2.5 he774522_0 conda-forge
zeromq 4.3.4 h0e60522_0 conda-forge
zipp 3.5.0 pyhd8ed1ab_0 conda-forge
zlib 1.2.11 h8ffe710_1013 conda-forge