2022-08-05 • Spiketrain correlations (of unconnected-but-detected)
Contents
2022-08-05 • Spiketrain correlations (of unconnected-but-detected)¶
Imports¶
#
using Revise
using MyToolbox
using VoltoMapSim
[ Info: Precompiling VoltoMapSim [f713100b-c48c-421a-b480-5fcb4c589a9e]
Params¶
Based on Roxin; same as previous nb’s.
d = 6
p = get_params(
duration = 10minutes,
p_conn = 0.04,
g_EE = 1 / d,
g_EI = 18 / d,
g_IE = 36 / d,
g_II = 31 / d,
ext_current = Normal(-0.5 * pA/√seconds, 5 * pA/√seconds),
E_inh = -80 * mV,
record_v = [1, 801],
);
Run sim¶
s = cached(sim, [p.sim]);
Running simulation: 100%|███████████████████████████████| Time: 0:11:16[39mmmmm
Saving output at `C:\Users\tfiers\.phdcache\datamodel v2 (net)\sim\ebefdc43edf54e31.jld2` … done (29.6 s)
s = augment_simdata(s, p);
Bin spiketrains¶
function bin(spiketimes; duration, binsize)
# `spiketimes` is assumed sorted.
# `duration` is of the spiketimes signal and determines the number of bins.
num_bins = ceil(Int, duration / binsize)
spikecounts = fill(0, num_bins)
# loop counters:
spike = 1
bin_end_time = binsize
for bin in 1:num_bins
while spiketimes[spike] < bin_end_time
spikecounts[bin] += 1
spike += 1
if spike > length(spiketimes)
return spikecounts
end
end
bin_end_time += binsize
end
end;
Test¶
events = s.spike_times[1][1:10]
show(events)
[0.685, 1.9, 2.8, 6.86, 8.55, 8.65, 13.4, 15.3, 16.9, 17.1]
show(bin(events; duration=20, binsize=2))
[2, 1, 0, 1, 2, 0, 1, 1, 2, 0]
Looks good
Apply¶
p.conntest.STA_window_length / ms
100
binned_spikes = [bin(s.spike_times[n], duration = 10minutes, binsize = 100ms) for n in s.neuron_IDs];
Correlation with recorded neuron¶
m = 1; # analyzed neuron
cors = [cor(binned_spikes[m], binned_spikes[n]) for n in s.neuron_IDs]; # Pearson corr
Split neurons by type¶
v = s.signals[m].v
ii = get_input_info(m, s, p);
ii.num_inputs
(exc = 26, inh = 10)
perf = cached(evaluate_conntest_perf, [v, ii.spiketrains, p], key = [p, m]);
Testing connections: 100%|██████████████████████████████| Time: 0:01:00
Saving output at `C:\Users\tfiers\.phdcache\datamodel v2 (net)\evaluate_conntest_perf\86c598a20b08f8eb.jld2` … done (4.3 s)
perf.detection_rates
(TPR_exc = 0.154, TPR_inh = 1, FPR = 0.15)
signif_unconn = ii.unconnected_neurons[findall(perf.p_values.unconn .< p.evaluation.α)];
tested_unconn = ii.unconnected_neurons[1:p.evaluation.N_tested_presyn]
insignif_unconn = [n for n in tested_unconn if n ∉ signif_unconn];
length(signif_unconn), length(insignif_unconn)
(6, 34)
Plot¶
using PyPlot
using VoltoMapSim.Plot
Plot.add_refline
add_refline (generic function with 1 method)
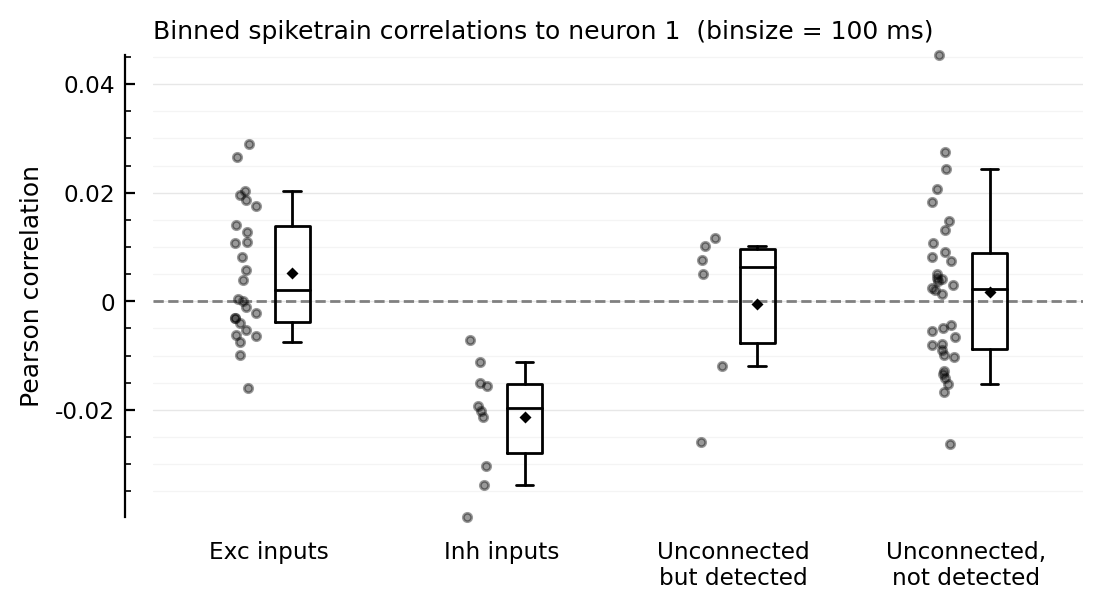
function plotcors(cors, binsize_ms)
ax = ydistplot(
"Exc inputs" => cors[ii.exc_inputs],
"Inh inputs" => cors[ii.inh_inputs],
"Unconnected\nbut detected" => cors[signif_unconn],
"Unconnected,\nnot detected" => cors[insignif_unconn],
figsize = (6, 3),
hylabel = "Binned spiketrain correlations to neuron $m (binsize = $(binsize_ms) ms)",
ylabel = "Pearson correlation",
# ylim = [-0.04, +0.045],
)
Plot.add_refline(ax, 0, zorder=1, c="gray")
end
plotcors(cors, 100);
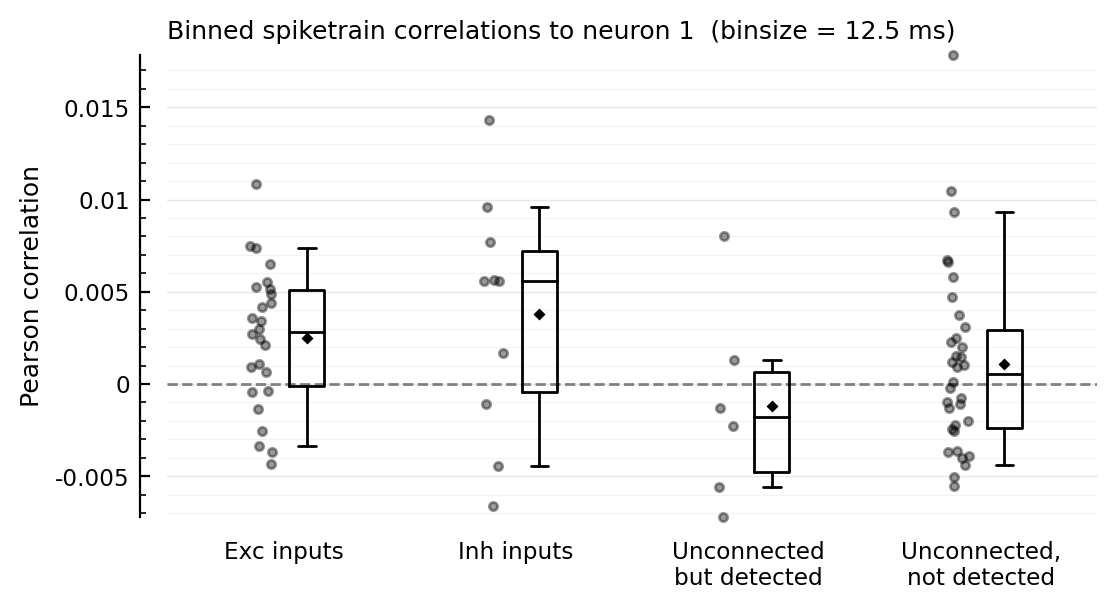
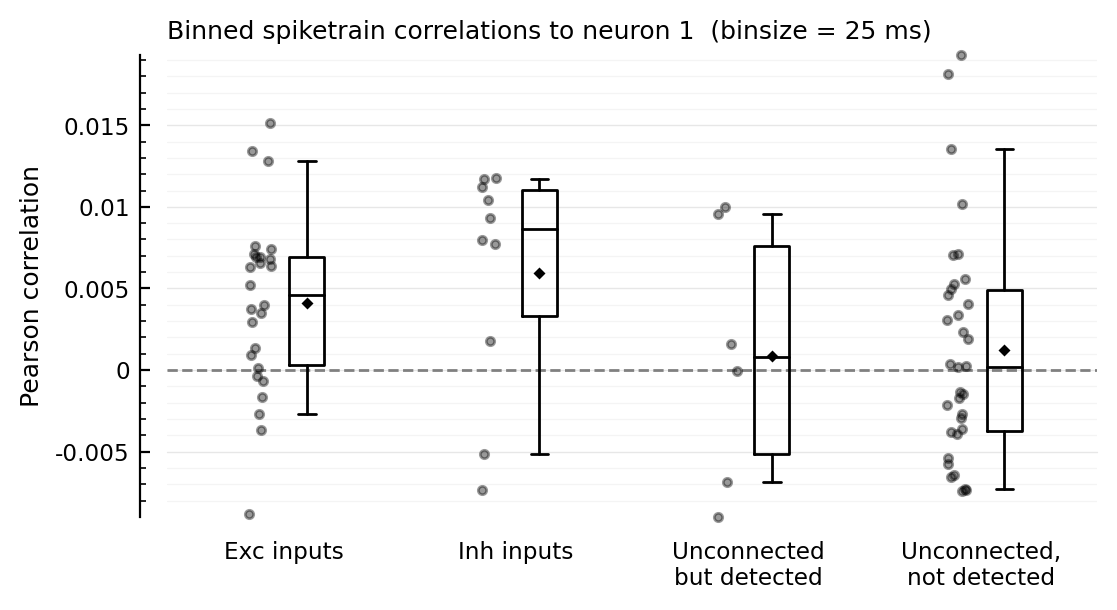
More bin sizes¶
function plotcors_for(; binsize)
binned_spikes = [bin(s.spike_times[n], duration = 10minutes; binsize) for n in s.neuron_IDs]
cors = [cor(binned_spikes[m], binned_spikes[n]) for n in s.neuron_IDs]
plotcors(cors, binsize/ms)
end;
for binsize in [12.5, 25, 50, 100, 200] * ms
plotcors_for(; binsize)
end;
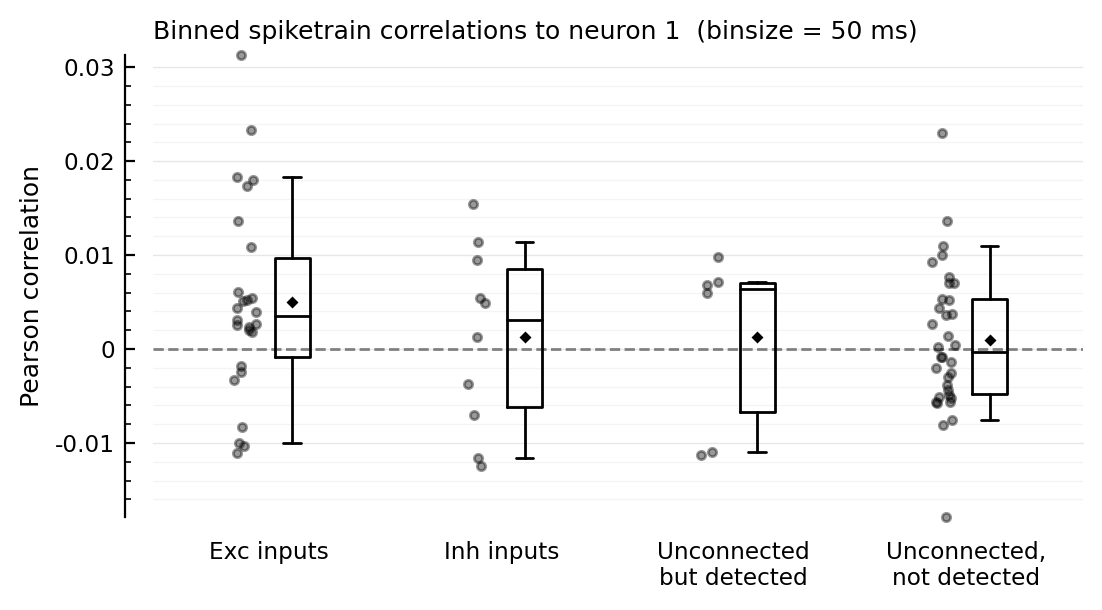
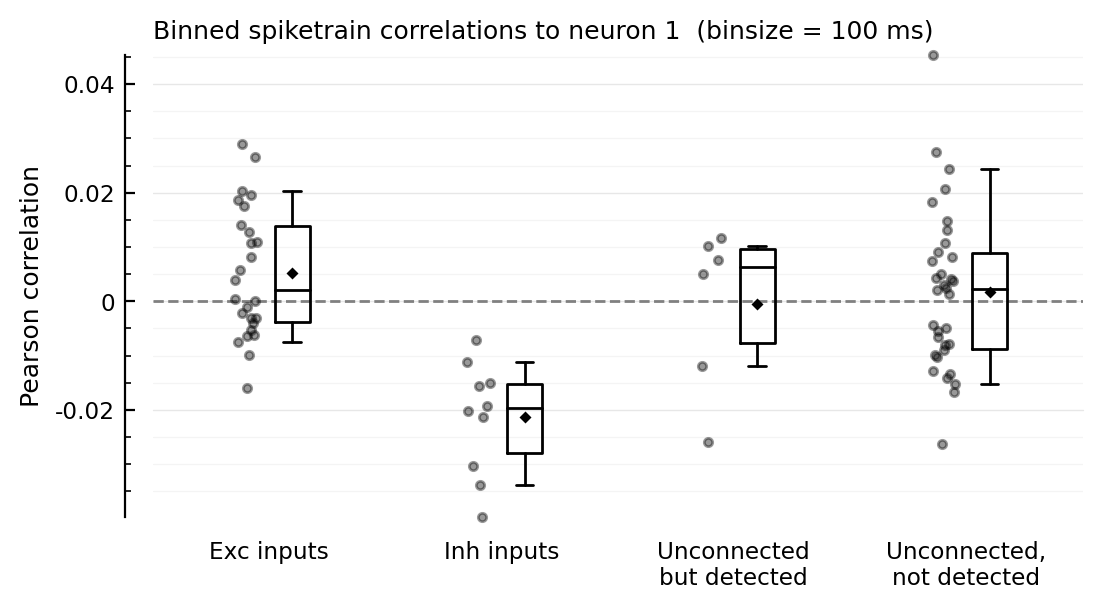
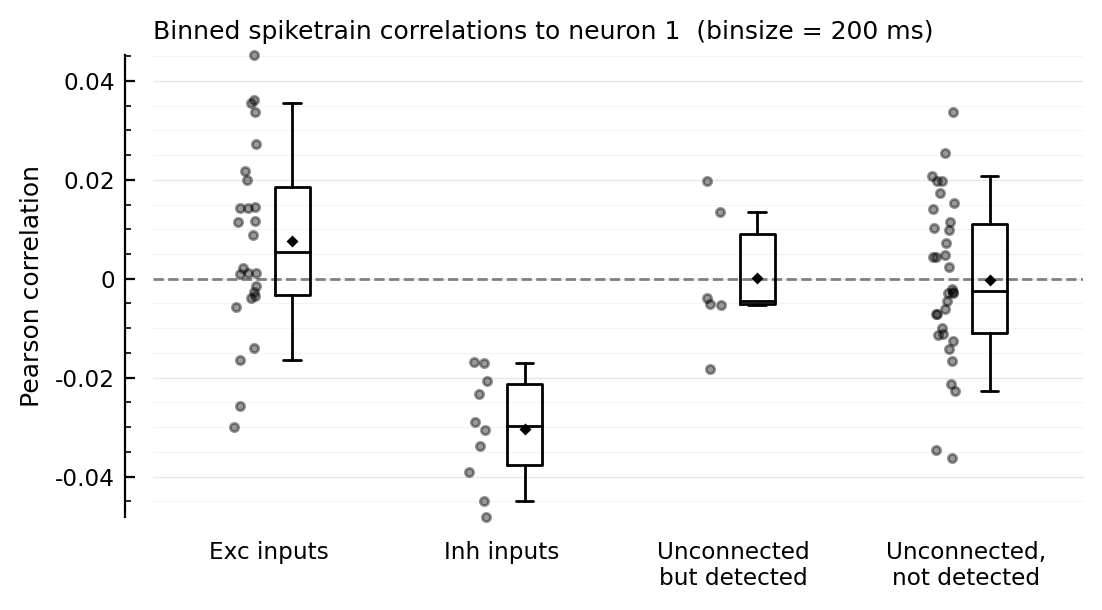
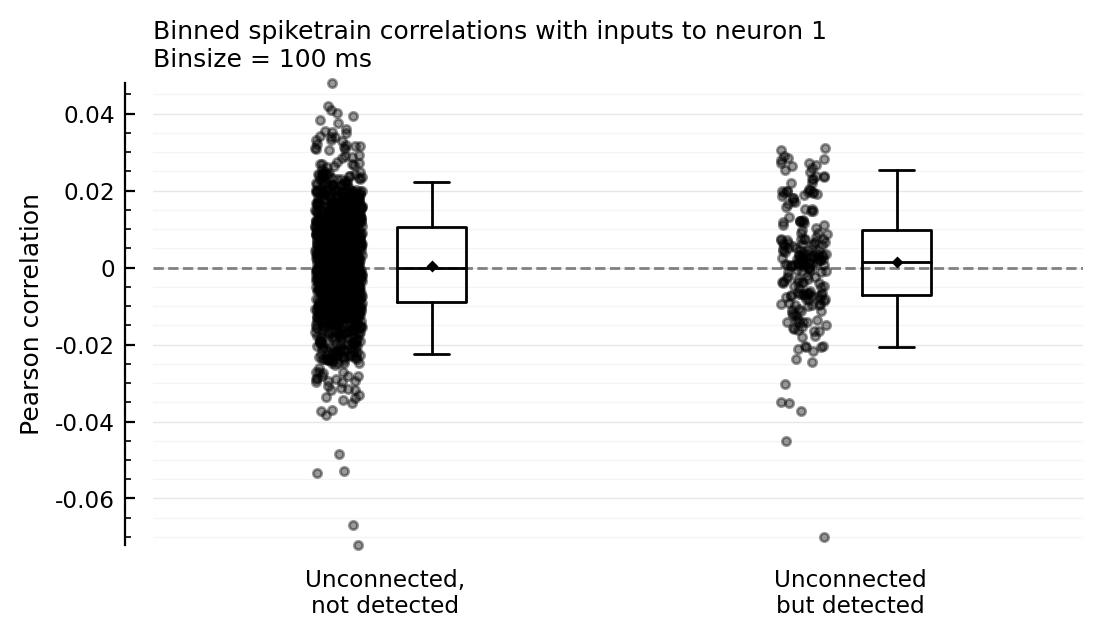
Correlation with other inputs¶
The guess: unconnected-but-detected are more correlated with actual inputs to the recorded neuron, than unconnected not detected are.
To test, we compute spiketrain correlations between inputs (and not between an input and the recorded neuron as above).
all_inputs = [ii.exc_inputs; ii.inh_inputs];
signif_unconn_cors = vcat(
[
[cor(binned_spikes[m], binned_spikes[n]) for n in all_inputs]
for m in signif_unconn
]...
)
insignif_unconn_cors = vcat(
[
[cor(binned_spikes[m], binned_spikes[n]) for n in all_inputs]
for m in insignif_unconn
]...
);
function plotcors_between_inputs(insignif_unconn_cors, signif_unconn_cors, binsize_ms = 100)
ax = ydistplot(
"Unconnected,\nnot detected" => insignif_unconn_cors,
"Unconnected\nbut detected" => signif_unconn_cors,
figsize = (6, 3),
hylabel = "Binned spiketrain correlations with inputs to neuron $m \nBinsize = $binsize_ms ms",
ylabel = "Pearson correlation",
# ylim = [-0.04, +0.045],
)
Plot.add_refline(ax, 0, zorder=1, c="gray")
end
plotcors_between_inputs(insignif_unconn_cors, signif_unconn_cors);
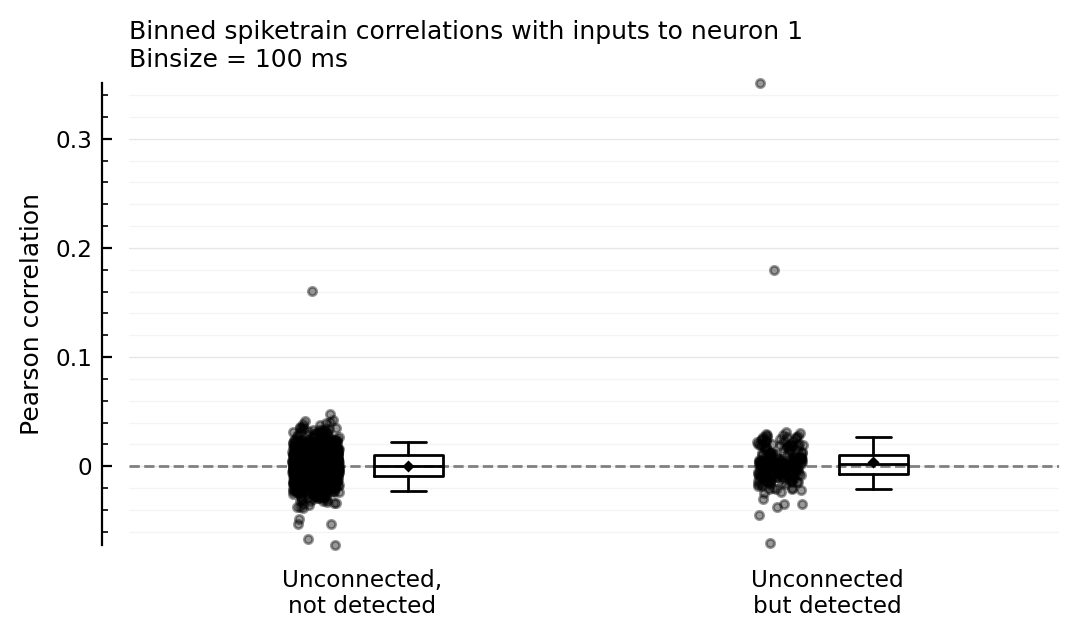
Remove outliers (manually) to zoom in
signif_unconn_cors__rm = signif_unconn_cors[signif_unconn_cors .< 0.1]
insignif_unconn_cors__rm = insignif_unconn_cors[insignif_unconn_cors .< 0.1]
plotcors_between_inputs(insignif_unconn_cors__rm, signif_unconn_cors__rm);