2022-09-15 • Two-pass connection test: ptp, then corr with found avg
Contents
2022-09-15 • Two-pass connection test: ptp, then corr with found avg¶
Two-pass connection test: peak-to-peak, then correlation with found average
Imports¶
#
using MyToolbox
using VoltoMapSim
[ Info: Precompiling VoltoMapSim [f713100b-c48c-421a-b480-5fcb4c589a9e]
Params¶
p = get_params(
duration = 10minutes,
p_conn = 0.04,
g_EE = 1,
g_EI = 1,
g_IE = 4,
g_II = 4,
ext_current = Normal(-0.5 * pA/√seconds, 5 * pA/√seconds),
E_inh = -80 * mV,
record_v = [1:40; 801:810],
);
Run sim¶
@time s = cached(sim, [p.sim]);
Loading cached output from `C:\Users\tfiers\.phdcache\datamodel v2 (net)\sim\ce5e30aee45561ed.jld2` … done (8.6 s)
9.992754 seconds (12.06 M allocations: 3.025 GiB, 9.21% gc time, 69.41% compilation time: 5% of which was recompilation)
@time s = augment(s, p);
2.605456 seconds (4.91 M allocations: 293.675 MiB, 8.31% gc time, 97.97% compilation time: 1% of which was recompilation)
1. peak-to-peak¶
We take all inputs with predicted_type :exc.
Current pval threshold is 0.05.
We could be stricter.
It’ll be tradeoff: stricter threshold gives less STAs to average to build our template; but there will be less noisy STAs mixed in (or even wrong STAs, i.e. of non-inputs).
Now
Get the :exc input detections for neuron 1
Average their STA
Apply this template to STAs of all inputs
# But first: cache STAs :)
Construct ‘tested_connections’ table¶
SimData = typeof(s);
function get_tested_connections(s::SimData, p::ExpParams)
# We do not test all N x N connections (that's too much).
# The connections we do test are determined by which neurons
# we record the voltage of, and the `N_tested_presyn` parameter.
tested_connections = DataFrame(
post = Int[], # neuron ID
pre = Int[], # neuron ID
conntype = Symbol[], # :exc or :inh or :unconn
posttype = Symbol[], # :exc or :inh
)
@unpack N_tested_presyn, rngseed = p.conntest
resetrng!(rngseed)
function get_labelled_sample(input_neurons, conntype)
# Example output: `[(3, :exc), (5, :exc), (12, :exc), …]`.
N = min(length(input_neurons), N_tested_presyn)
inputs_sample = sample(input_neurons, N, replace = false, ordered = true)
zip(inputs_sample, fill(conntype, N))
end
recorded_neurons = keys(s.v) |> collect |> sort!
for post in recorded_neurons
posttype = s.neuron_type[post]
inputs_to_test = chain(
get_labelled_sample(s.exc_inputs[post], :exc),
get_labelled_sample(s.inh_inputs[post], :inh),
get_labelled_sample(s.non_inputs[post], :unconn),
) |> collect
for (pre, conntype) in inputs_to_test
push!(tested_connections, (; post, pre, conntype, posttype))
end
end
return tested_connections
end
tc = get_tested_connections(s, p)
disp(tc, 3)
3,906 rows × 4 columns
| post | pre | conntype | posttype | |
|---|---|---|---|---|
| Int64 | Int64 | Symbol | Symbol | |
| 1 | 1 | 139 | exc | exc |
| 2 | 1 | 681 | exc | exc |
| 3 | 1 | 11 | exc | exc |
| ⋮ | ⋮ | ⋮ | ⋮ | ⋮ |
Calc all STAs¶
using Base.Threads
function calc_all_STAs(s::SimData, p::ExpParams)
# Multi-threaded calculation of the real and shuffled STAs of all tested connections.
#
# We use a Channel to gather calculations,
# as inserting items into a Dict is not thread-safe.
@unpack rngseed, num_shuffles = p.conntest
tc = tested_connections = get_tested_connections(s, p)
recorded_neurons = unique(tested_connections.post)
ch = Channel(Inf) # `Inf` size, so no blocking on insert
@info "Using $(nthreads()) threads"
pbar = Progress(nrow(tested_connections), desc = "Calculating STAs: ")
@threads(
for m in recorded_neurons
v = s.v[m]
inputs_to_test = tc.pre[tc.post .== m]
for n in inputs_to_test
spikes = s.spike_times[n]
real_STA = calc_STA(v, spikes, p)
shuffled_STAz = Vector(undef, num_shuffles) # [1]
resetrng!(rngseed)
for i in 1:num_shuffles
shuffled_spikes = shuffle_ISIs(spikes)
shuffled_STAz[i] = calc_STA(v, shuffled_spikes, p)
end
put!(ch, ((n => m), real_STA, shuffled_STAz))
next!(pbar)
end
end)
# Empty the channel, into the output dicts
STAs = Dict{Pair{Int, Int}, Vector{Float64}}()
shuffled_STAs = Dict{Pair{Int, Int}, Vector{Vector{Float64}}}()
while !isempty(ch)
conn, real, shuffleds = take!(ch)
STAs[conn] = real
shuffled_STAs[conn] = shuffleds
end
close(ch)
return (; tested_connections, STAs, shuffled_STAs)
end
# [1] Giving this the same name as the output dict, in combination with `@threads`,
# confuses type inference and makes it infer `Any` for the `shuffled_STAs` output dict.
# (That doesn't matter though, as this is a user-facing, top-level function; not an inner-loop one).
calc_all_STAs (generic function with 1 method)
# For 4000 connections tested (0.4% of all) and 100 shuffles,
# we get 404_000 STAs.
# At 1000 Float64 samples per STA, that is 3.2 GB.
# On typing (code_warntype) of calc_all_STAs:
# it didn't matter for timing it was untyped
# (tested with small params `q`, 447 conns).
out = cached(calc_all_STAs, [s,p], key=[p.sim, p.conntest]);
[ Info: Using 7 threads
Calculating STAs: 100%|█████████████████████████████████| Time: 0:19:04
Saving output at `C:\Users\tfiers\.phdcache\datamodel v2 (net)\calc_all_STAs\b9353bdd11d8b8cb.jld2` … done (18.2 s)
@time (out = cached(calc_all_STAs, [s,p], key=[p.sim, p.conntest]));
Loading cached output from `C:\Users\tfiers\.phdcache\datamodel v2 (net)\calc_all_STAs\b9353bdd11d8b8cb.jld2` … done (19.3 s)
19.725458 seconds (9.35 M allocations: 3.348 GiB, 28.71% gc time, 10.98% compilation time)
tested_connections, STAs, shuffled_STAs = out;
Calc pval¶
Sig = Vector{Float64}
function calc_pval(real_STA::Sig, shuffled_STAs::Vector{Sig}, test_stat)
real_t = test_stat(real_STA)
shuffled_ts = [test_stat(STA) for STA in shuffled_STAs]
num_shuffled_larger = count(shuffled_ts .> real_t)
N = length(shuffled_STAs)
return if num_shuffled_larger == 0
(pval = 1/N,
pval_type = "<")
else
(pval = num_shuffled_larger / N,
pval_type = "=")
end
end
calc_pval (generic function with 1 method)
Test connections¶
function test_conn__ptp((from,to), STAs, shuffled_STAs; α)
# α is the p-value threshold.
pval, _ = calc_pval(STAs[from=>to], shuffled_STAs[from=>to], ptp)
area = area_over_start(STAs[from=>to])
if (pval > α) predtype = :unconn
elseif (area > 0) predtype = :exc
else predtype = :inh end
return (; predtype, pval, area)
end;
test_conn__ptp((from,to); α) = test_conn__ptp((from,to), STAs, shuffled_STAs; α);
test_conn__ptp(139=>1, α = 0.01)
(predtype = :exc, pval = 0.01, area = 0.16991151700597443)
testresults =
@showprogress "Testing connections: " (
map(eachrow(tested_connections)) do r
test_conn__ptp(r.pre => r.post, α = 0.01)
end
);
Testing connections: 100%|██████████████████████████████| Time: 0:00:04
tc = hcat(tested_connections, DataFrame(testresults))
select!(tc, :posttype, :post, :pre, :conntype, :predtype, :pval, :area)
disp(tc, 3)
3,906 rows × 7 columns
| posttype | post | pre | conntype | predtype | pval | area | |
|---|---|---|---|---|---|---|---|
| Symbol | Int64 | Int64 | Symbol | Symbol | Float64 | Float64 | |
| 1 | exc | 1 | 139 | exc | exc | 0.01 | 0.169912 |
| 2 | exc | 1 | 681 | exc | unconn | 0.02 | -0.0399586 |
| 3 | exc | 1 | 11 | exc | exc | 0.01 | 0.180399 |
| ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ |
(“predtype” is predicted conntype).
We used strict α of 0.01.
Out of interest, what is the T and FPR.
Detection rates¶
Previously, we had detection rates per recorded (‘post’) neuron. We can also just calculate the rates over the whole table. Corresponds to the mean of previous notebooks.
detrate(typ) = count((tc.conntype .== typ) .& (tc.predtype .== typ)) / count(tc.conntype .== typ)
(
TPR_exc = detrate(:exc),
TPR_inh = detrate(:inh),
FPR = 1 - detrate(:unconn),
)
(TPR_exc = 0.521, TPR_inh = 0.831, FPR = 0.0785)
We’ll use E→E connections for our template.
How many of these did we detect,
and how many of the detected are false.
# so we search for [posttype=:exc],
# and then - [conntype=:exc, predtpye=:exc]
# - [conntype=:unconn, predtype=:exc]
This is precision, not TPR (aka accuracy).
Precision¶
using DataFramesMeta
df = tc[tc.posttype .== :exc, :]
precision = count((df.predtype .== :exc) .& (df.conntype .== :exc)) / count(df.predtype .== :exc)
0.925
num_EE_detections = count(df.predtype .== :exc)
692
correct_EE_detections = precision * num_EE_detections
640
incorrect_EE_detections = (1 - precision) * num_EE_detections
52
Note that this presumes we know the type of each neuron (‘post’).
Yeah that’s not general.
Let’s instead just take all exc detected connections, whether or not post is exc or inh.
..of all detected exc connections¶
(So also exc→inh)
# sub back df > tc
precision = count((tc.predtype .== :exc) .& (tc.conntype .== :exc)) / count(tc.predtype .== :exc)
0.934
num_exc_detections = count(tc.predtype .== :exc)
845
correct_exc_detections = precision * num_exc_detections
789
incorrect_exc_detections = (1 - precision) * num_exc_detections
56
So to summarize, we detected 52% of true exc connections,
and 93% of what we detected as exc is actually exc.
2. STA template¶
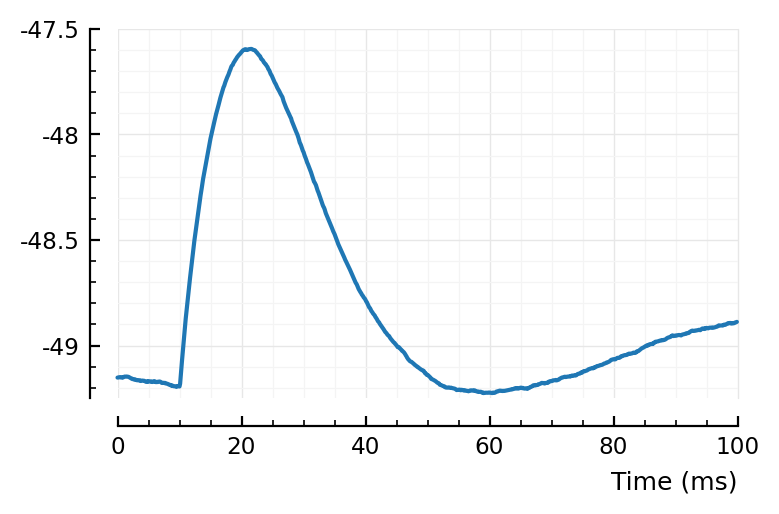
det_exc_conns = [r.pre=>r.post for r in eachrow(tc) if r.predtype == :exc];
template_ptp = mean(STAs[c] for c in det_exc_conns);
plotsig(template_ptp / mV, p);
# now to corr this, for all conns.
function test_conn__corr((from,to), STAs, shuffled_STAs; α, template)
# α is the p-value threshold.
STA = STAs[from=>to]
corr = cor(STA, template)
if (corr > 0) test_stat = (STA -> cor(STA, template))
else test_stat = (STA -> -cor(STA, template)) end
pval, _ = calc_pval(STA, shuffled_STAs[from=>to], test_stat)
if (pval > α) predtype = :unconn
elseif (corr > 0) predtype = :exc
else predtype = :inh end
return (; predtype, pval, corr)
end
# note: closure of `template`
test_conn__corr((from,to) ; α, template = template_ptp) =
test_conn__corr((from,to), STAs, shuffled_STAs; α, template);
function test_conns(testfunc; α)
testresults =
@showprogress "Testing connections: " (
map(eachrow(tested_connections)) do r
testfunc(r.pre => r.post; α)
end)
tc = hcat(tested_connections, DataFrame(testresults))
# Reorder first cols:
select!(tc, :posttype, :post, :pre, :conntype, :)
num_TP(typ) = count((tc.conntype .== typ) .& (tc.predtype .== typ))
num_real(typ) = count(tc.conntype .== typ)
num_detected(typ) = count(tc.predtype .== typ)
TPR(typ) = num_TP(typ) / num_real(typ)
precision(typ) = num_TP(typ) / num_detected(typ)
perf = (
TPR_exc = TPR(:exc),
TPR_inh = TPR(:inh),
FPR = 1 - TPR(:unconn),
prec_exc = precision(:exc),
prec_inh = precision(:inh),
prec_unconn = precision(:unconn),
)
return (; tc, perf)
end
res_2pass = test_conns(test_conn__corr, α = 0.05);
Testing connections: 100%|██████████████████████████████| Time: 0:00:01
Note α = 5% here; in the ptp step, it was the stricter 1%.
res_2pass.perf
(TPR_exc = 0.8151815181518152, TPR_inh = 0.9360613810741688, FPR = 0.14800000000000002, prec_exc = 0.8988355167394468, prec_inh = 0.6918714555765595, prec_unconn = 0.8507239141288068)
(Note that TPR no longer broken down by postsynaptic type here. Also, we added precision as performance measure: how much of detected are correct).
Compare performance¶
..with just ptp-and-area, now with α = 0.05 too:
res_ptp = test_conns(test_conn__ptp, α = 0.05);
Testing connections: 100%|██████████████████████████████| Time: 0:00:01
To compare with the previous correlation-test-notebook, where we cheated by using the average STA of all a-priori known E→E connections as template, ..
..we used different measure there (calculate detection rates per postsynaptic neuron, and then take the median of that. Also these rates were broken down by postsyn type). The measures here are simpler.
EE_conns = [r.pre=>r.post for r in eachrow(tc) if (r.conntype == :exc) & (r.posttype .== :exc)];
avg_EE_STA = mean(STAs[c] for c in EE_conns);
plotsig(zscore(avg_EE_STA), p, label = "real avg E→E STA");
plotsig(zscore(template_ptp), p, label = "avg exc STA from ptp test", hylabel="z-scored")
plt.legend();
# zscore, as ptp template has larger range (hm!)
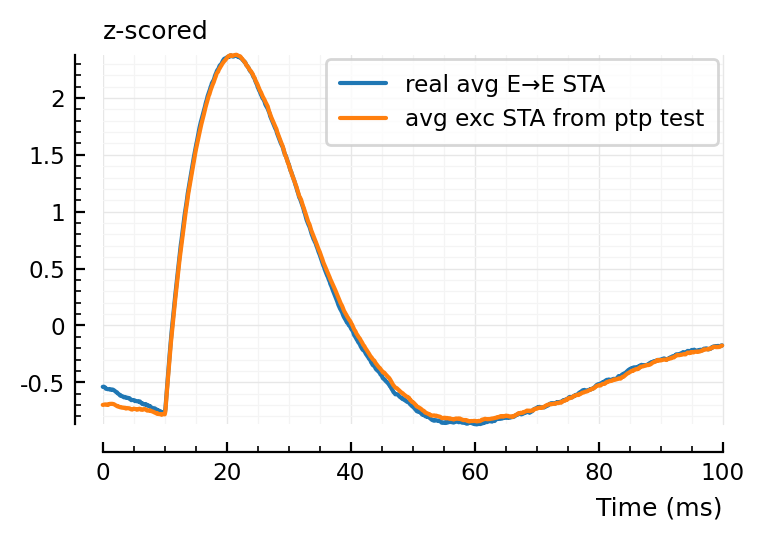
We should not calc the real E→E STA to compare,
but rather the E→any STA.
exc_conns = [r.pre=>r.post for r in eachrow(tc) if (r.conntype == :exc)];
avg_exc_STA = mean(STAs[c] for c in exc_conns);
plotsig(zscore(avg_exc_STA), p, label = "real avg exc STA");
plotsig(zscore(template_ptp), p, label = "avg exc STA from ptp test", hylabel="z-scored")
plt.legend();
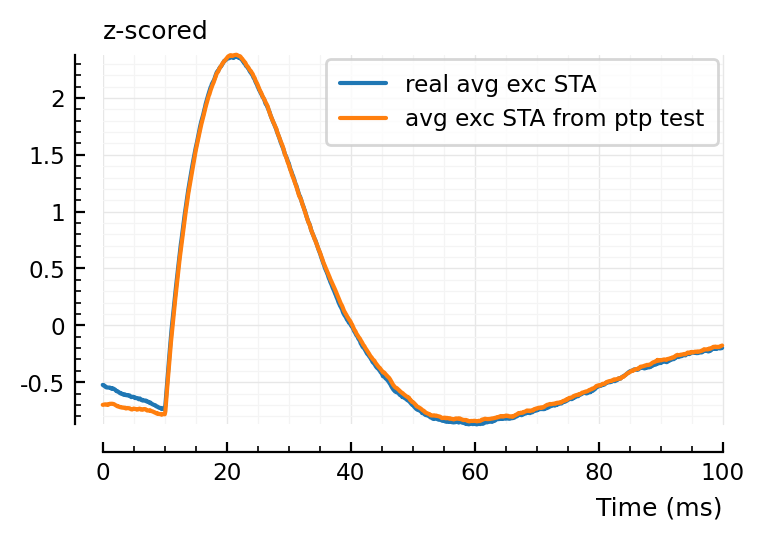
Not a large difference it seems (not identical either).
Also interesting that the ptp STA has a larger range than the real avg STA:
plotsig(avg_exc_STA / mV, p, label = "real avg exc STA");
plotsig(template_ptp / mV, p, label = "avg exc STA from ptp test", hylabel="mV")
plt.legend();
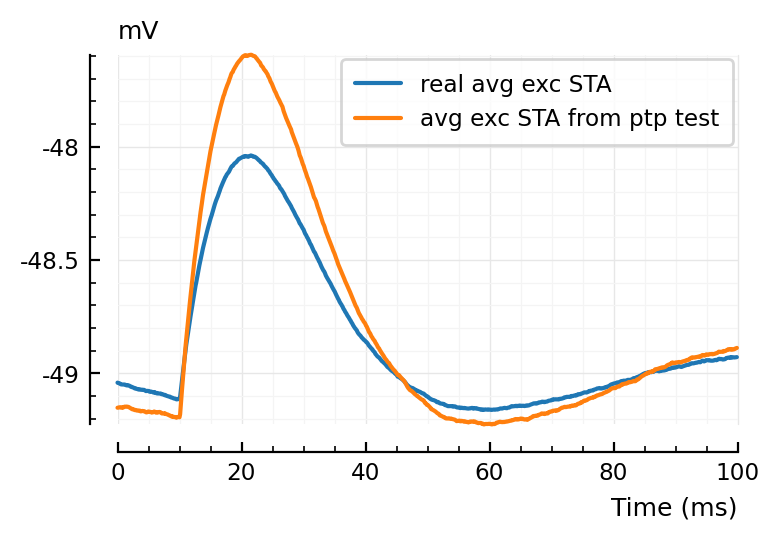
Ah of course: reason why real average STA is smaller, is cause it also includes weaker connections, which the strict ptp test did not pick up.
# Now use this real avg STA as template
testfunc(conn; α) = test_conn__corr(conn; α, template = avg_exc_STA);
res_real_avg = test_conns(testfunc, α = 0.05);
Testing connections: 100%|██████████████████████████████| Time: 0:00:00
res_real_avg.perf
(TPR_exc = 0.8145214521452145, TPR_inh = 0.9360613810741688, FPR = 0.14700000000000002, prec_exc = 0.8981077147016011, prec_inh = 0.6958174904942965, prec_unconn = 0.8504486540378864)
df = DataFrame([
(; method="ptp only", res_ptp.perf...),
(; method="ptp-then-corr", res_2pass.perf...),
(; method="corr w/ real avg", res_real_avg.perf...),
]
);
Summary table¶
printsimple(df)
method TPR_exc TPR_inh FPR prec_exc prec_inh prec_unconn
────────────────────────────────────────────────────────────────────────────
ptp only 0.574 0.849 0.145 0.889 0.466 0.772
ptp-then-corr 0.815 0.936 0.148 0.899 0.692 0.851
corr w/ real avg 0.815 0.936 0.147 0.898 0.696 0.85
So the two-pass connection test idea works well – very nearly as well as the ‘cheating’ test where we correlate with the real average excitatory STA (instead of an average STA found after a first-pass, strict peak-to-peak test).
On what the numbers mean:
FPR is how many non-connections were classified as connected.
85% precision_unconn means that 15% of what we said was not connected actually was.
By also calculating precision (aka positive predictive value) as a performance measure, we see that our high inhibitory detection rates trade-off against a lower precision, compared to excitatory connections.
Confusion matrix¶
..with counts, and ratios too.
function calc_perf_measures(tc::DataFrame)
# `tc` is a `tested_connections` table with `predtype` (prediction) column added.
conntypes = [:unconn, :exc, :inh]
conntypes_matrix = [(pred, real) for pred in conntypes, real in conntypes]
counts = similar(conntypes_matrix, Int) # Detection counts
for (i, (pred, real)) in enumerate(conntypes_matrix)
counts[i] = count((tc.predtype .== pred) .& (tc.conntype .== real))
end
N = length(conntypes)
sensitivities = Vector(undef, N) # aka TPRs
precisions = Vector(undef, N)
for i in 1:N
num_correct = counts[i,i]
num_real = sum(counts[:,i])
num_predicted = sum(counts[i,:])
sensitivities[i] = num_correct / num_real
precisions[i] = num_correct / num_predicted
end
return (; counts, sensitivities, precisions, conntypes, conntypes_matrix)
end;
function make_perf_display(tested_connections::DataFrame)
data = calc_perf_measures(tested_connections)
titlerow = titlecol = 1
grouprow = groupcol = 2
datarows = datacols = 3:5
sens_row = prec_col = 7
nrows = ncols = 7
cells = Matrix{Any}(undef, nrows, ncols)
fill!(cells, "")
cells[grouprow, datacols] .= data.conntypes
cells[datarows, groupcol] .= data.conntypes
cells[datarows, datacols] .= data.counts
cells[datarows, titlecol] .= [
" ┌",
"Predicted type",
" └",
]
cells[titlerow, datacols] .= ["┌───────", "Real type", "───────┐"]
fmt_pct(x) = join([round(Int, 100x), "%"])
cells[sens_row, titlecol] = "Sensitivity"
cells[sens_row, datacols] .= fmt_pct.(data.sensitivities)
cells[titlerow, prec_col] = "Precision"
cells[datarows, prec_col] .= fmt_pct.(data.precisions)
title = join(["Tested connections: ", sum(data.counts)])
bold_cells = vcat([(titlerow,c) for c in 1:ncols], [(r,titlecol) for r in 1:nrows])
return DisplayTable(cells, title, bold_cells)
end;
(I made displaytable.jl in ../pkg/MyToolbox/src/)
t = make_perf_display(res_2pass.tc);
print(t)
Tested connections: 3906
┌─────── Real type ───────┐ Precision
unconn exc inh
┌ unconn 1704 274 25 85%
Predicted type exc 139 1235 0 90%
└ inh 157 6 366 69%
Sensitivity 85% 82% 94%
Show¶
display(t)
| Tested connections: 3906 | ||||||
|---|---|---|---|---|---|---|
| ┌─────── | Real type | ───────┐ | Precision | |||
unconn | exc | inh | ||||
| ┌ | unconn | 1704 | 274 | 25 | 85% | |
| Predicted type | exc | 139 | 1235 | 0 | 90% | |
| └ | inh | 157 | 6 | 366 | 69% | |
| Sensitivity | 85% | 82% | 94% |
The above is for the two-pass method (strict ptp, then corr with found average exc STA).
Below is for the ptp-only method.
Both at α = 5%.
make_perf_display(res_ptp.tc)
| Tested connections: 3906 | ||||||
|---|---|---|---|---|---|---|
| ┌─────── | Real type | ───────┐ | Precision | |||
unconn | exc | inh | ||||
| ┌ | unconn | 1709 | 459 | 47 | 77% | |
| Predicted type | exc | 97 | 870 | 12 | 89% | |
| └ | inh | 194 | 186 | 332 | 47% | |
| Sensitivity | 85% | 57% | 85% |
(Sensitivity = TPR).