2021-07-30 • Inhibitory connections
Contents
2021-07-30 • Inhibitory connections¶
Hello back. We’re re-running the simulation as before, only this time ‘stimulating’ our one Izhikevich neuron with inhibitory spike trains; that is: we’ll lower the reversal potential of the synapse.
What we did previously: aggregate all incoming spikes together (in N_to_1_experiment.simulate), then use this one spike train to calculate a conductance signal (exp decay; in synapses.py); then feed this conductance signal to sim_izh, where a synaptic current I_syn is calculated as g_syn[i] * (v[i] - v_syn), with v_syn a fixed synaptic reversal potential.
We’ll need to change this architecture:
We have to calculate a conductance signal for every incoming spike train separately (we can keep
synapses.pyas is).sim_izhneeds to take not just oneg_synsignal; but one for every spike train.v_synis no longer a property of the neuron; it is a property of every incoming spike train.
Our new simulation code in the codebase will no longer work in older notebooks (but that’s why we have git and the repro info).
Where to start? What is our eventual goal here?
Well the question is if we can still detect connections from activities alone. We did it with STA height previously. Won’t work now right?
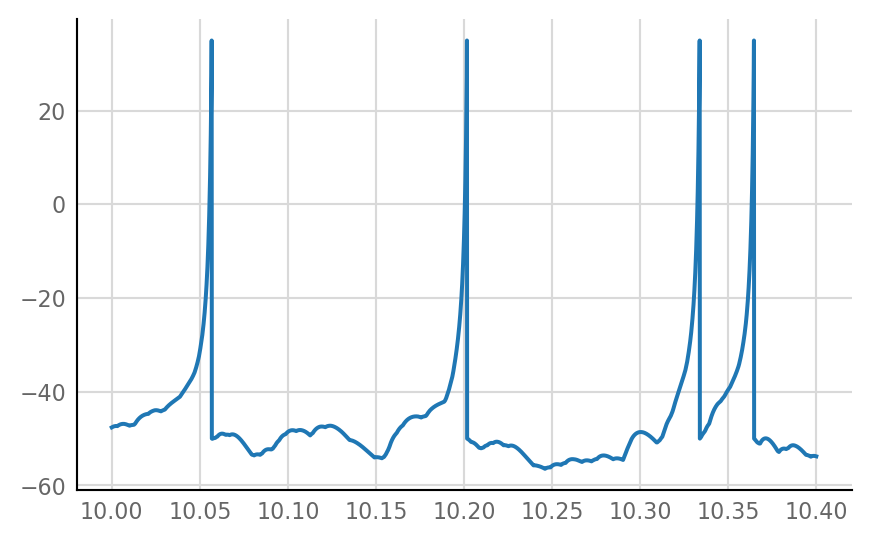
Let’s start by plotting izh output (v_mem) and STA with a negative v_syn.
Prelude¶
from voltage_to_wiring_sim.notebook_init import *
Preloading: numpy, numba, matplotlib.pyplot, seaborn.
Importing from submodules … ✔
Imported `np`, `mpl`, `plt`, `sns`, `pd`
Imported codebase (`voltage_to_wiring_sim`) as `v`
Imported `*` from `v.support.units`
Setup autoreload
v.print_reproducibility_info()
This cell was last run by tfiers on yoga on Mon 23 Aug 2021, at 22:28 (UTC+0200).
Last git commit (Mon 23 Aug 2021, 19:05).
Uncommited changes to 1 file.
Neuron input¶
Here we generate a bunch of spike trains, each paired with a synaptic reversal potential.
v_syn_E = 0 * mV
v_syn_I = -50 * mV;
We choose the inhibitory reversal potential to be 10 mV higher than the resting potential. This is what’s found e.g. here for GABA neurons: https://www.nature.com/articles/ncomms1377.pdf
num_spike_trains = 30
p_inhibitory = 0.33
p_connected = 0.6
spike_rate = 20 * Hz;
Other parameters, same as in previous notebooks:
from voltage_to_wiring_sim.sim.izhikevich_neuron import cortical_RS
sim_duration = 10 * minute
timestep = 0.1 * ms
spike_rate = 20 * Hz
Δg_syn = 0.8 * nS
τ_syn = 7 * ms
neuron_params = cortical_RS
imaging_spike_SNR = 20;
Now for the actual input generation.
from voltage_to_wiring_sim.sim.poisson_spikes import generate_Poisson_spikes
from random import random
input_spike_trains = np.empty(num_spike_trains, dtype=object)
v_syn = np.empty(num_spike_trains, dtype=float)
is_connected = np.zeros(num_spike_trains, dtype=bool)
for i in range(num_spike_trains):
input_spike_trains[i] = generate_Poisson_spikes(spike_rate, sim_duration)
if random() < p_connected:
is_connected[i] = True
if random() < p_inhibitory:
v_syn[i] = v_syn_I
else:
v_syn[i] = v_syn_E
connected_and_E = indices_where(is_connected & (v_syn == v_syn_E))
connected_and_I = indices_where(is_connected & (v_syn == v_syn_I))
not_connected = indices_where(~is_connected);
Izhikevich simulation¶
I edited the sim_izh to take a list of v_syns and g_syn signals.
Let’s make the g_syn signals.
from voltage_to_wiring_sim.sim.synapses import calc_synaptic_conductance
def indices_where(bool_array):
return np.nonzero(bool_array)[0]
g_syn = []
for i in indices_where(is_connected):
g_syn.append(calc_synaptic_conductance(sim_duration, timestep, input_spike_trains[i], Δg_syn, τ_syn))
v_syn_connected = v_syn[indices_where(is_connected)];
..and now pass them to the izh sim code.
from voltage_to_wiring_sim.sim.izhikevich_neuron import simulate_izh_neuron
izh_output = simulate_izh_neuron(sim_duration, timestep, neuron_params, v_syn_connected, g_syn);
from voltage_to_wiring_sim.support.signal import plot_signal
plot_slice = dict(t_start=10 * second, duration=400 * ms);
plot_signal(izh_output.V_m.slice(**plot_slice) / mV);
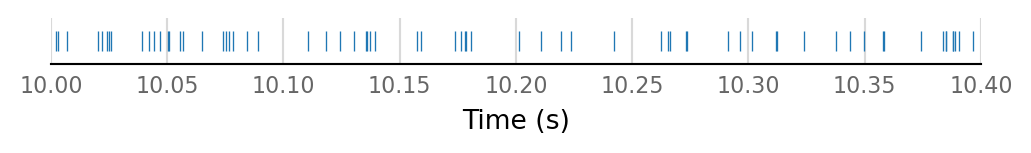
all_connected_I_spikes = np.concatenate([st for st in input_spike_trains[connected_and_I]])
v.support.spike_train.plot_spike_train(all_connected_I_spikes, [10, 10.4]);
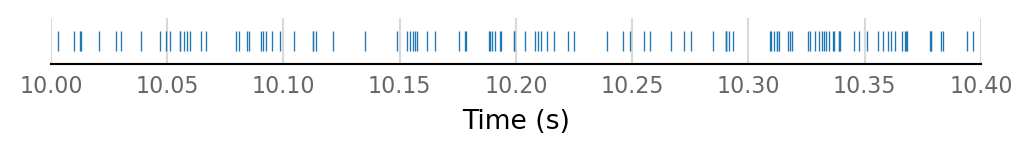
all_connected_E_spikes = np.concatenate([st for st in input_spike_trains[connected_and_E]])
v.support.spike_train.plot_spike_train(all_connected_E_spikes, [10, 10.4]);
VI noise & STAs¶
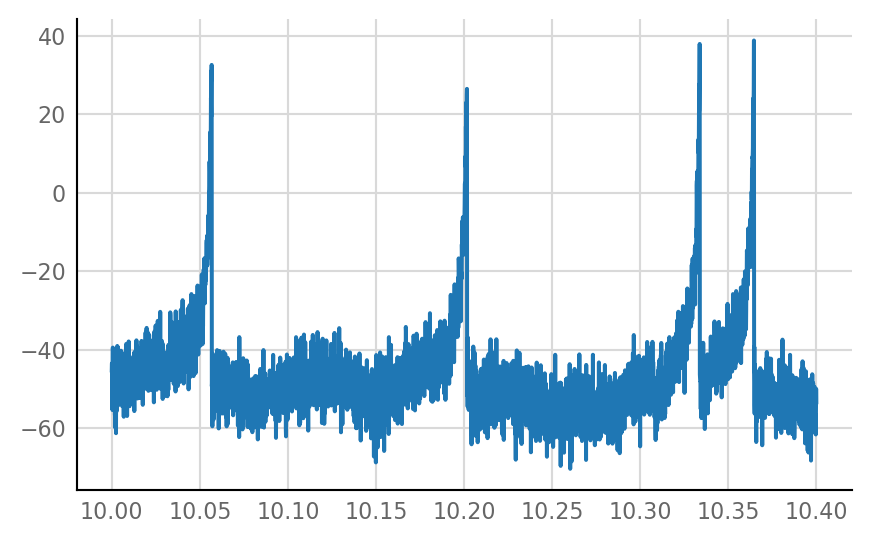
VI_signal = v.sim.imaging.add_VI_noise(izh_output.V_m, neuron_params, imaging_spike_SNR);
plot_signal(VI_signal.slice(**plot_slice) / mV);
window_duration = 100 * ms;
def plot_STA(spike_train_index):
STA = v.conntest.STA.calculate_STA(VI_signal, input_spike_trains[spike_train_index], window_duration)
ax = plot_signal(STA / mV, time_units=ms)
# ax.set_ylim([-53, -50]) # mV
ax.set_ylim([-48.5, -45]) # mV
Results¶
No shuffle test for now; just visually inspecting the STAs.
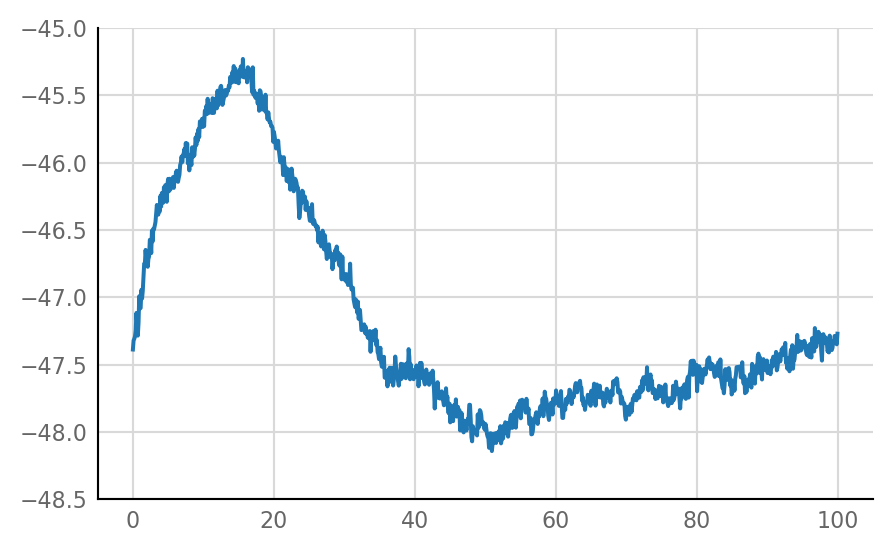
Connected excitatory and unconnected are as before:
plot_STA(connected_and_E[0])
plot_STA(not_connected[0])
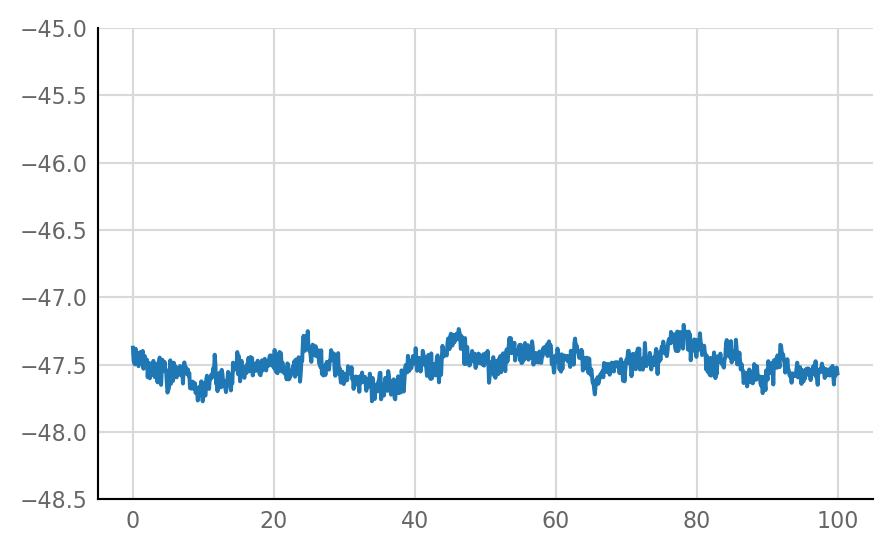
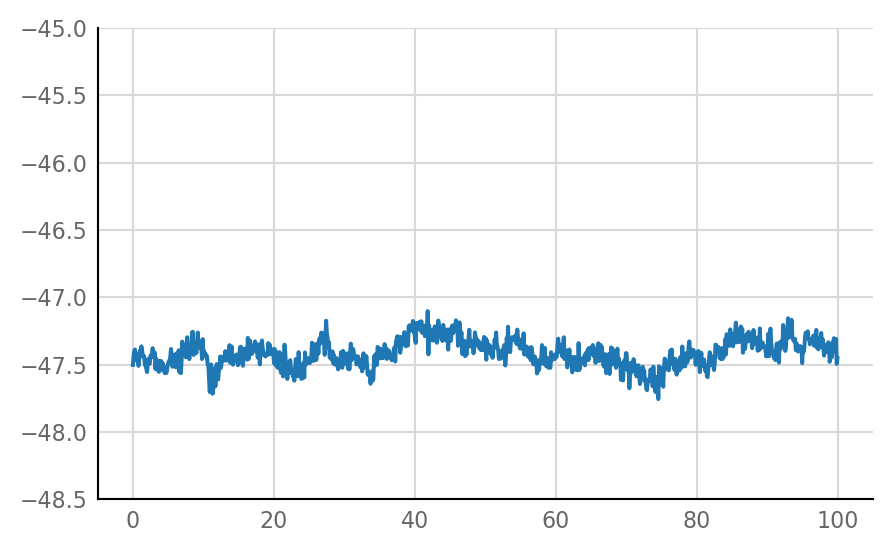
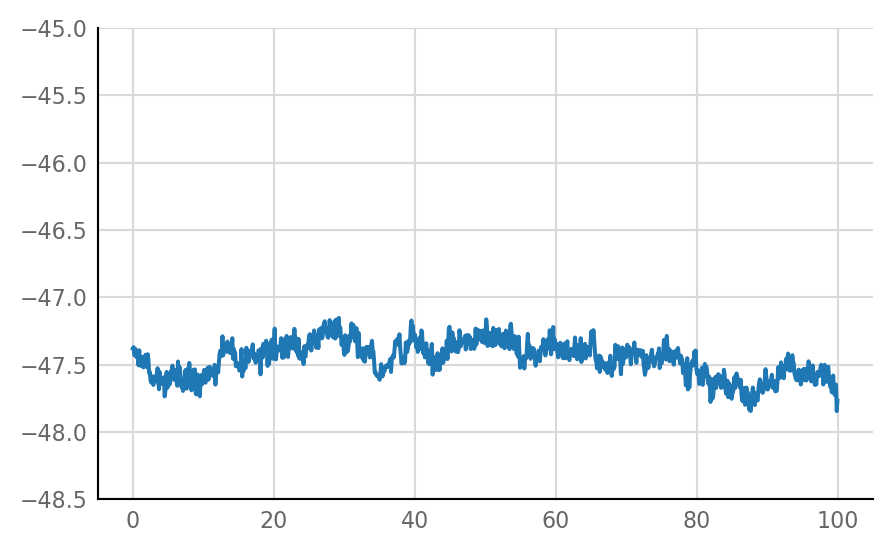
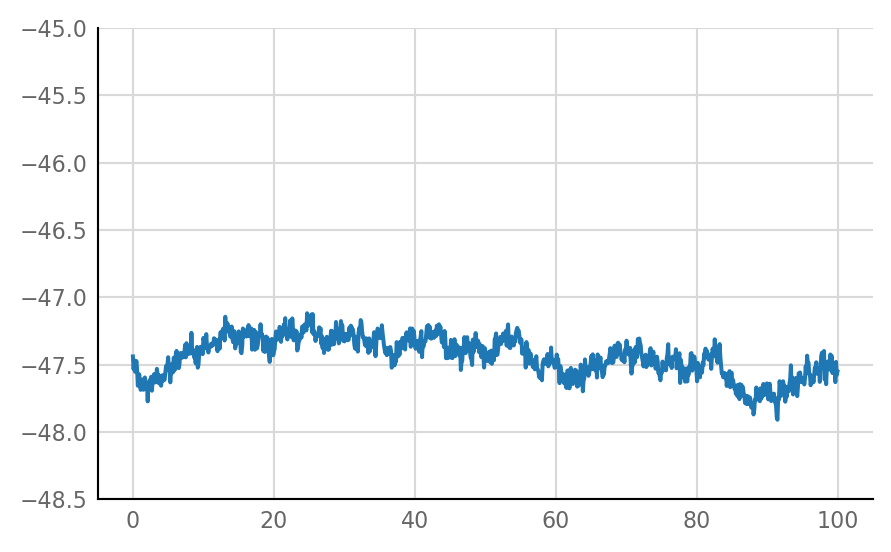
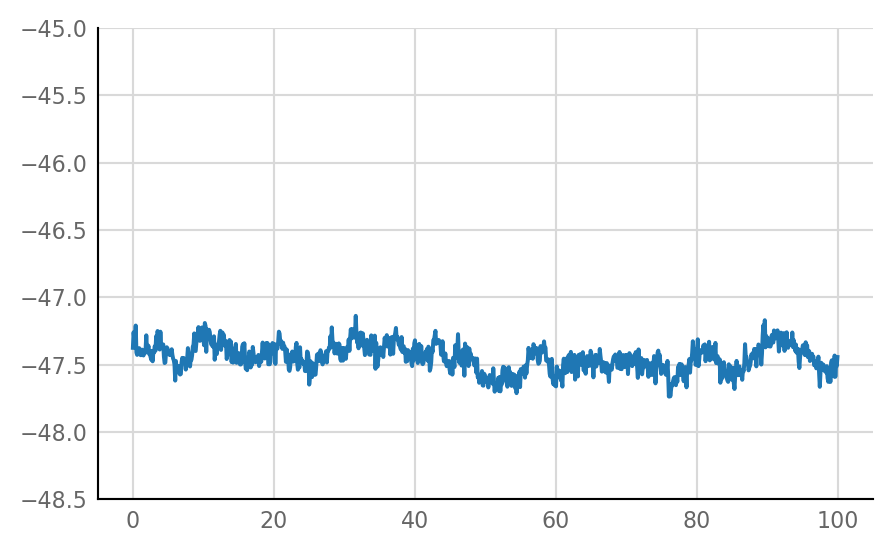
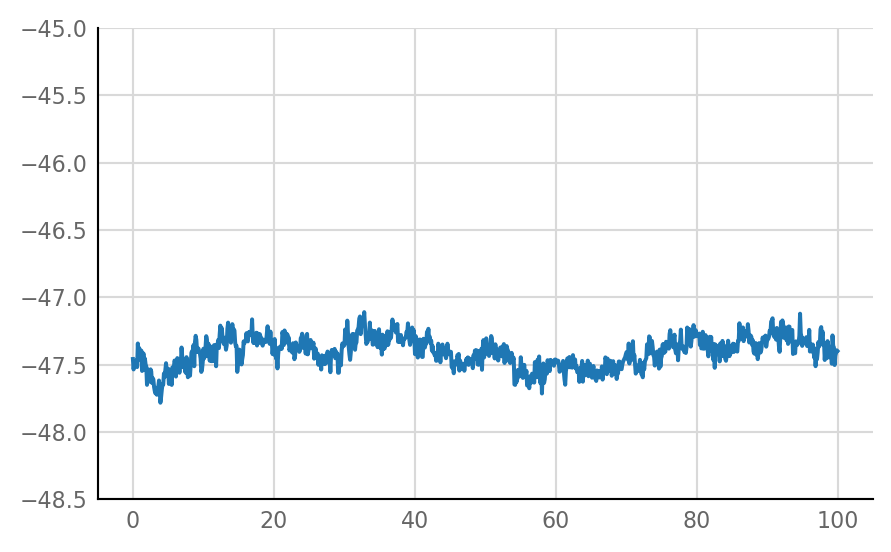
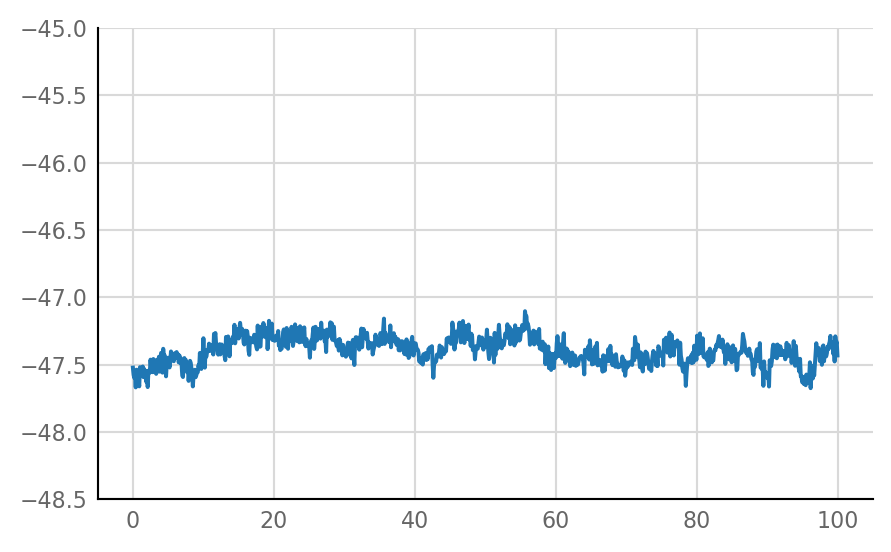
Inhibitory STA does not look much different from unconnected (i.e. same as shuffled) STA, alas:
for i in range(len(connected_and_I)):
plot_STA(connected_and_I[i])
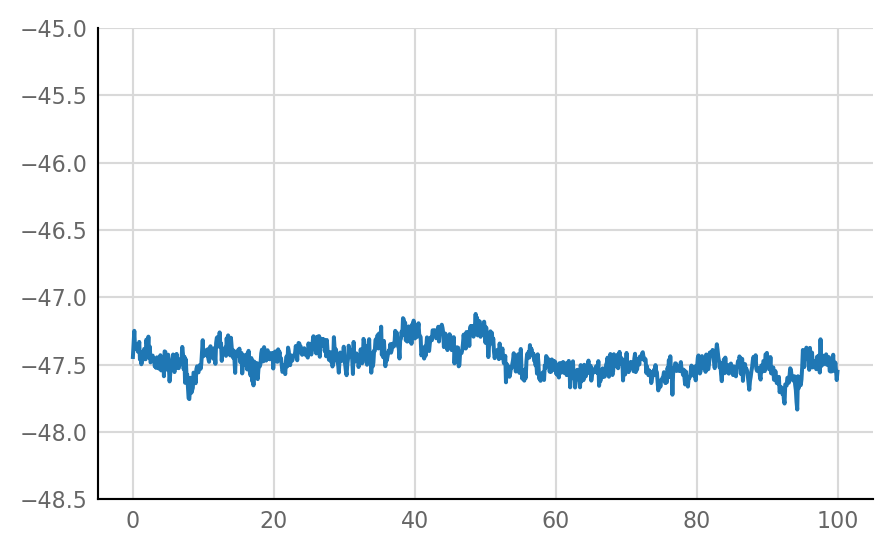
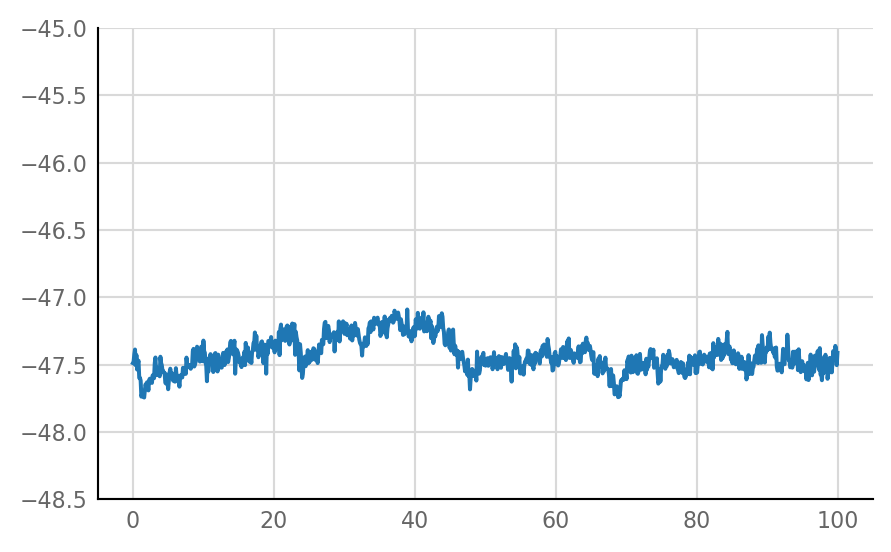
Reproducibility¶
v.print_reproducibility_info(verbose=True)
This cell was last run by tfiers on yoga
on Mon 23 Aug 2021, at 18:13 (UTC+0200).
Last git commit (Fri 20 Aug 2021, 21:41).
Uncommited changes to:
M codebase/tests/test_run_embedded_tests.py
M codebase/voltage_to_wiring_sim/N_to_1_experiment.py
M codebase/voltage_to_wiring_sim/__init__.py
M codebase/voltage_to_wiring_sim/sim/__init__.py
M codebase/voltage_to_wiring_sim/sim/izhikevich_neuron.py
M codebase/voltage_to_wiring_sim/sim/neuron_params.py
RM codebase/voltage_to_wiring_sim/sim/input_spike_trains.py -> codebase/voltage_to_wiring_sim/sim/poisson_spikes.py
M codebase/voltage_to_wiring_sim/sim/synapses.py
M notebooks/2021-07-30__inhibitory.ipynb
Platform:
Windows-10
CPython 3.8.6 (C:\conda\python.exe)
Intel(R) Core(TM) i7-10510U CPU @ 1.80GHz
Dependencies of voltage_to_wiring_sim and their installed versions:
numpy 1.19.5
matplotlib 3.3.4
numba 0.52.0
joblib 1.0.1
seaborn 0.11.1
scipy 1.6.0
preload 2.2
nptyping 1.4.0
Full conda list:
# packages in environment at C:\conda:
#
# Name Version Build Channel
anyio 2.0.2 py38haa244fe_4 conda-forge
appdirs 1.4.4 pyh9f0ad1d_0 conda-forge
argon2-cffi 20.1.0 py38h294d835_2 conda-forge
async_generator 1.10 py_0 conda-forge
atomicwrites 1.4.0 pyh9f0ad1d_0 conda-forge
attrs 20.3.0 pyhd3deb0d_0 conda-forge
babel 2.9.0 pyhd3deb0d_0 conda-forge
backcall 0.2.0 pyh9f0ad1d_0 conda-forge
backports 1.0 py_2 conda-forge
backports.functools_lru_cache 1.6.1 py_0 conda-forge
black 20.8b1 py_1 conda-forge
bleach 3.3.0 pyh44b312d_0 conda-forge
bokeh 2.2.3 py38haa244fe_0 conda-forge
brotlipy 0.7.0 py38h294d835_1001 conda-forge
bzip2 1.0.8 h8ffe710_4 conda-forge
ca-certificates 2020.12.5 h5b45459_0 conda-forge
certifi 2020.12.5 py38haa244fe_1 conda-forge
cffi 1.14.4 py38hd8c33c5_1 conda-forge
chardet 4.0.0 py38haa244fe_1 conda-forge
click 7.1.2 pyh9f0ad1d_0 conda-forge
cloudpickle 1.6.0 py_0 conda-forge
colorama 0.4.4 pyh9f0ad1d_0 conda-forge
conda 4.9.2 py38haa244fe_0 conda-forge
conda-package-handling 1.7.2 py38h8934438_0 conda-forge
console_shortcut 0.1.1 4
cryptography 3.3.1 py38hd8c33c5_1 conda-forge
cycler 0.10.0 py_2 conda-forge
cytoolz 0.11.0 py38h294d835_3 conda-forge
dask 2021.1.0 pyhd8ed1ab_0 conda-forge
dask-core 2021.1.0 pyhd8ed1ab_0 conda-forge
dataclasses 0.7 pyhb2cacf7_7 conda-forge
decorator 4.4.2 py_0 conda-forge
defusedxml 0.6.0 py_0 conda-forge
distributed 2021.1.1 py38haa244fe_0 conda-forge
docutils 0.16 pypi_0 pypi
entrypoints 0.3 pyhd8ed1ab_1003 conda-forge
freetype 2.10.4 h546665d_1 conda-forge
fsspec 0.8.5 pyhd8ed1ab_0 conda-forge
gitdb 4.0.5 pyhd8ed1ab_1 conda-forge
gitpython 3.1.12 pyhd8ed1ab_0 conda-forge
heapdict 1.0.1 py_0 conda-forge
icu 68.1 h0e60522_0 conda-forge
idna 2.10 pyh9f0ad1d_0 conda-forge
importlib-metadata 3.4.0 py38haa244fe_0 conda-forge
importlib_metadata 3.4.0 hd8ed1ab_0 conda-forge
iniconfig 1.1.1 pyh9f0ad1d_0 conda-forge
intel-openmp 2020.3 h57928b3_311 conda-forge
ipykernel 5.4.2 py38h7b7c402_0 conda-forge
ipython 7.20.0 py38hc5df569_1 conda-forge
ipython_genutils 0.2.0 py_1 conda-forge
ipywidgets 7.6.3 pyhd3deb0d_0 conda-forge
jedi 0.18.0 py38haa244fe_2 conda-forge
jinja2 2.11.3 pyh44b312d_0 conda-forge
joblib 1.0.1 pypi_0 pypi
jpeg 9d h8ffe710_0 conda-forge
json5 0.9.5 pyh9f0ad1d_0 conda-forge
jsonschema 3.2.0 py_2 conda-forge
jupyter_client 6.1.11 pyhd8ed1ab_1 conda-forge
jupyter_contrib_core 0.3.3 py_2 conda-forge
jupyter_contrib_nbextensions 0.5.1 py38h32f6830_1 conda-forge
jupyter_core 4.7.1 py38haa244fe_0 conda-forge
jupyter_highlight_selected_word 0.2.0 py38haa244fe_1002 conda-forge
jupyter_latex_envs 1.4.6 py38h32f6830_1001 conda-forge
jupyter_nbextensions_configurator 0.4.1 py38haa244fe_2 conda-forge
jupyter_server 1.2.2 py38haa244fe_1 conda-forge
jupyterlab 3.0.5 pyhd8ed1ab_0 conda-forge
jupyterlab-classic 0.1.3 pyhd8ed1ab_0 conda-forge
jupyterlab-sublime 0.4.1 pypi_0 pypi
jupyterlab_pygments 0.1.2 pyh9f0ad1d_0 conda-forge
jupyterlab_server 2.1.4 pyhd8ed1ab_0 conda-forge
jupyterlab_widgets 1.0.0 pyhd8ed1ab_1 conda-forge
jupytext 1.10.0 pyh44b312d_0 conda-forge
jupytray 0.7.post10+dirty dev_0 <develop>
keyring 21.8.0 pypi_0 pypi
kiwisolver 1.3.1 py38hbd9d945_1 conda-forge
krb5 1.17.2 hbae68bd_0 conda-forge
libarchive 3.5.1 h8686738_1 conda-forge
libblas 3.9.0 8_mkl conda-forge
libcblas 3.9.0 8_mkl conda-forge
libclang 11.0.1 default_h5c34c98_1 conda-forge
libcurl 7.71.1 h4b64cdc_8 conda-forge
libiconv 1.16 he774522_0 conda-forge
liblapack 3.9.0 8_mkl conda-forge
libpng 1.6.37 h1d00b33_2 conda-forge
libsodium 1.0.18 h8d14728_1 conda-forge
libsolv 0.7.17 h7755175_0 conda-forge
libssh2 1.9.0 hb06d900_5 conda-forge
libtiff 4.2.0 hc10be44_0 conda-forge
libxml2 2.9.10 hf5bbc77_3 conda-forge
libxslt 1.1.33 h65864e5_2 conda-forge
llvmlite 0.35.0 py38h57a6900_1 conda-forge
locket 0.2.0 py_2 conda-forge
lxml 4.6.2 py38h292cb97_1 conda-forge
lz4-c 1.9.3 h8ffe710_0 conda-forge
lzo 2.10 he774522_1000 conda-forge
m2w64-gcc-libgfortran 5.3.0 6 conda-forge
m2w64-gcc-libs 5.3.0 7 conda-forge
m2w64-gcc-libs-core 5.3.0 7 conda-forge
m2w64-gmp 6.1.0 2 conda-forge
m2w64-libwinpthread-git 5.0.0.4634.697f757 2 conda-forge
mamba 0.7.8 py38hdd88130_0 conda-forge
markdown-it-py 0.6.2 pyhd8ed1ab_0 conda-forge
markdown2 2.4.0 pypi_0 pypi
markupsafe 1.1.1 py38h294d835_3 conda-forge
matplotlib-base 3.3.4 py38h34ddff4_0 conda-forge
mdit-py-plugins 0.2.5 pyhd8ed1ab_0 conda-forge
menuinst 1.4.16 py38he774522_1
mistune 0.8.4 py38h294d835_1003 conda-forge
mkl 2020.4 hb70f87d_311 conda-forge
more-itertools 8.6.0 pyhd8ed1ab_0 conda-forge
mpmath 1.1.0 py_0 conda-forge
msgpack-python 1.0.2 py38hbd9d945_1 conda-forge
msys2-conda-epoch 20160418 1 conda-forge
multipledispatch 0.6.0 pypi_0 pypi
multitasking 0.0.9 pypi_0 pypi
mypy_extensions 0.4.3 py38haa244fe_3 conda-forge
natsort 7.1.1 pyhd8ed1ab_0 conda-forge
nb_conda_kernels 2.3.1 py38haa244fe_0 conda-forge
nbclassic 0.2.6 pyhd8ed1ab_0 conda-forge
nbclient 0.5.1 py_0 conda-forge
nbconvert 6.0.7 py38haa244fe_3 conda-forge
nbdime 2.1.0 py_0 conda-forge
nbformat 5.0.8 py_0 conda-forge
nest-asyncio 1.4.3 pyhd8ed1ab_0 conda-forge
notebook 6.2.0 py38haa244fe_0 conda-forge
nptyping 1.4.0 pypi_0 pypi
numba 0.52.0 py38h4c96930_0 conda-forge
numpy 1.19.5 py38h0cc643e_1 conda-forge
oauthlib 3.1.0 pypi_0 pypi
oboe 0.2 pypi_0 pypi
olefile 0.46 pyh9f0ad1d_1 conda-forge
openssl 1.1.1i h8ffe710_0 conda-forge
packaging 20.8 pyhd3deb0d_0 conda-forge
pandas 1.2.1 py38h4c96930_0 conda-forge
pandas-flavor 0.2.0 py_0 conda-forge
pandoc 2.11.4 h8ffe710_0 conda-forge
pandocfilters 1.4.2 py_1 conda-forge
parso 0.8.1 pyhd8ed1ab_0 conda-forge
partd 1.1.0 py_0 conda-forge
pathspec 0.8.1 pyhd3deb0d_0 conda-forge
patsy 0.5.1 py_0 conda-forge
pickleshare 0.7.5 py_1003 conda-forge
pillow 8.1.0 py38hf7ce48b_1 conda-forge
pip 21.0.1 pyhd8ed1ab_0 conda-forge
pkginfo 1.6.1 pypi_0 pypi
pluggy 0.13.1 py38haa244fe_4 conda-forge
powershell_shortcut 0.0.1 3
preload 2.2 pypi_0 pypi
prometheus_client 0.9.0 pyhd3deb0d_0 conda-forge
prompt-toolkit 3.0.14 pyha770c72_0 conda-forge
psutil 5.8.0 py38h294d835_1 conda-forge
puprelease 1.8.1 dev_0 <develop>
py 1.10.0 pyhd3deb0d_0 conda-forge
pycosat 0.6.3 py38h294d835_1006 conda-forge
pycparser 2.20 pyh9f0ad1d_2 conda-forge
pygments 2.7.4 pyhd8ed1ab_0 conda-forge
pyopenssl 20.0.1 pyhd8ed1ab_0 conda-forge
pypandoc 1.6.3 pypi_0 pypi
pyparsing 2.4.7 pyh9f0ad1d_0 conda-forge
pyqt 5.12.3 py38haa244fe_7 conda-forge
pyqt-impl 5.12.3 py38h885f38d_7 conda-forge
pyqt5-sip 4.19.18 py38h885f38d_7 conda-forge
pyqtchart 5.12 py38h885f38d_7 conda-forge
pyqtwebengine 5.12.1 py38h885f38d_7 conda-forge
pyrsistent 0.17.3 py38h294d835_2 conda-forge
pysocks 1.7.1 py38haa244fe_3 conda-forge
pytest 6.2.1 py38haa244fe_1 conda-forge
python 3.8.6 h7840368_4_cpython conda-forge
python-dateutil 2.8.1 py_0 conda-forge
python_abi 3.8 1_cp38 conda-forge
pytz 2021.1 pyhd8ed1ab_0 conda-forge
pywin32 300 py38h294d835_0 conda-forge
pywin32-ctypes 0.2.0 pypi_0 pypi
pywinpty 0.5.7 py38h32f6830_1 conda-forge
pyyaml 5.4.1 py38h294d835_0 conda-forge
pyzmq 22.0.1 py38h7a0e47e_0 conda-forge
qt 5.12.9 h5909a2a_3 conda-forge
readme-renderer 28.0 pypi_0 pypi
regex 2020.11.13 py38h294d835_1 conda-forge
reproc 14.2.1 h8ffe710_0 conda-forge
reproc-cpp 14.2.1 h0e60522_0 conda-forge
requests 2.25.1 pyhd3deb0d_0 conda-forge
requests-oauthlib 1.3.0 pypi_0 pypi
requests-toolbelt 0.9.1 pypi_0 pypi
rfc3986 1.4.0 pypi_0 pypi
ruamel_yaml 0.15.80 py38h294d835_1004 conda-forge
scikit-learn 0.24.1 py38ha09990b_0 conda-forge
scipy 1.6.0 py38h5f893b4_0 conda-forge
seaborn 0.11.1 h57928b3_0 conda-forge
seaborn-base 0.11.1 pyhd8ed1ab_1 conda-forge
send2trash 1.5.0 py_0 conda-forge
setuptools 49.6.0 py38haa244fe_3 conda-forge
setuptools-scm 5.0.1 pypi_0 pypi
six 1.15.0 pyh9f0ad1d_0 conda-forge
smmap 3.0.5 pyh44b312d_0 conda-forge
snakeviz 2.1.0 pyh9f0ad1d_0 conda-forge
sniffio 1.2.0 py38haa244fe_1 conda-forge
sortedcontainers 2.3.0 pyhd8ed1ab_0 conda-forge
sqlite 3.34.0 h8ffe710_0 conda-forge
statsmodels 0.12.2 py38h347fdf6_0 conda-forge
sympy 1.7.1 py38haa244fe_1 conda-forge
tblib 1.6.0 py_0 conda-forge
terminado 0.9.2 py38haa244fe_0 conda-forge
testpath 0.4.4 py_0 conda-forge
tfiers 0.3.2 dev_0 <develop>
threadpoolctl 2.1.0 pyh5ca1d4c_0 conda-forge
tk 8.6.10 h8ffe710_1 conda-forge
toml 0.10.2 pyhd8ed1ab_0 conda-forge
toolz 0.11.1 py_0 conda-forge
tornado 6.1 py38h294d835_1 conda-forge
tqdm 4.56.0 pyhd8ed1ab_0 conda-forge
traitlets 5.0.5 py_0 conda-forge
tweepy 3.10.0 pypi_0 pypi
twine 3.3.0 pypi_0 pypi
typed-ast 1.4.2 py38h294d835_0 conda-forge
typing_extensions 3.7.4.3 py_0 conda-forge
typish 1.9.1 pypi_0 pypi
unitlib 0.3 pypi_0 pypi
unyt 2.8.0 pypi_0 pypi
urllib3 1.26.3 pyhd8ed1ab_0 conda-forge
vc 14.2 hb210afc_2 conda-forge
voltage-to-wiring-sim 0.1 dev_0 <develop>
vs2015_runtime 14.28.29325 h5e1d092_0 conda-forge
wcwidth 0.2.5 pyh9f0ad1d_2 conda-forge
webencodings 0.5.1 py_1 conda-forge
wheel 0.36.2 pyhd3deb0d_0 conda-forge
widgetsnbextension 3.5.1 py38haa244fe_4 conda-forge
win_inet_pton 1.1.0 py38haa244fe_2 conda-forge
wincertstore 0.2 py38haa244fe_1006 conda-forge
windows-curses 2.2.0 pypi_0 pypi
winpty 0.4.3 4 conda-forge
winshell 0.6 pypi_0 pypi
xarray 0.16.2 pyhd8ed1ab_0 conda-forge
xlrd 2.0.1 pyhd8ed1ab_3 conda-forge
xz 5.2.5 h62dcd97_1 conda-forge
yaml 0.2.5 he774522_0 conda-forge
yfinance 0.1.55 pypi_0 pypi
zeromq 4.3.3 h0e60522_3 conda-forge
zict 2.0.0 py_0 conda-forge
zipp 3.4.0 py_0 conda-forge
zlib 1.2.11 h62dcd97_1010 conda-forge
zstd 1.4.8 h4e2f164_1 conda-forge