2020-07-06 • Single neuron simulation
Contents
2020-07-06 • Single neuron simulation¶
Introduction¶
I want to test whether we can recover connections..
between simulated neurons
under ‘realistic’ voltage imaging conditions
using spike-triggered averages of subhtreshold voltages.
Here we start by simulating a single neuron excited by an incoming spike train.
Izhikevich neuron¶
Model and parameters from Humphries 2006, “Understanding and using Izhikevich’s simple model neuron”.
Integration by forward Euler.
Parameters for a cortical regular spiking (RS) neuron.
Units are pF, mV, ms, and pA.
(a is in 1/ms).
C = 100; k = 0.7; v_r = -60; v_t = -40; v_peak = 35; a = 0.03; b = -2; c = -50; d = 100;
T = 1000 # simulation duration
dt = 0.1 # timestep
N = round(T/dt) # number of simulation steps
import numpy as np
from numba import jit
@jit
def izh_neuron(I):
'''
Input I and output v: arrays of length N.
'''
dv_dt = lambda v,u,i: (k*(v-v_r)*(v-v_t) - u + i)/C
du_dt = lambda v,u: a*(b*(v-v_r) - u)
v_t0 = v_r
u_t0 = 0
v = np.ones(N) * v_t0
u = np.ones(N) * u_t0
for i in range(N-1):
v[i+1] = v[i] + dt * dv_dt(v[i], u[i], I[i])
u[i+1] = u[i] + dt * du_dt(v[i], u[i])
if v[i+1] >= v_peak:
v[i] = v_peak
v[i+1] = c
u[i+1] = u[i+1] + d
return v
Plotting function we’ll reuse a few times.
import matplotlib.pyplot as plt
def plot_signal(x, ylabel='', t=None):
fig, ax = plt.subplots()
if t is None:
N = len(x)
T = N*dt
t = np.linspace(0, T, N)
ax.plot(t, x);
ax.set(xlabel="t (ms)", ylabel=ylabel)
return fig, ax
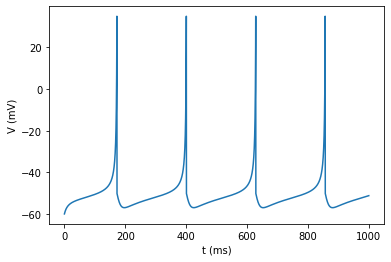
Test neuron model with constant current injection.
v = izh_neuron(I=60*np.ones(N));
plot_signal(v, 'V (mV)');
Presynaptic spikes¶
We use the same approach as in eg Dayan & Abott to generate (approximate) Poisson spike times.
(Approximate Poisson because we ignore the possibility of a neuron spiking more than once in the same small timebin dt).
f_spike = 1/1000 # Hz/1000
from numpy.random import seed, random
seed(0)
def spikes():
spikes = np.zeros(N)
for i in range(N):
spikes[i] = f_spike * dt > random()
return spikes
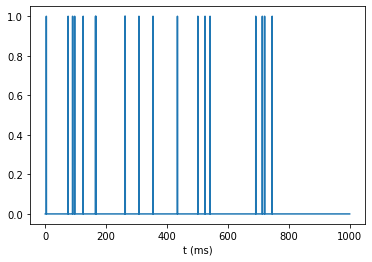
Aggregate spikes for all incoming neurons
N_in = 20
all_spikes = sum([spikes() for incoming_neuron in range(N_in)])
plot_signal(all_spikes);
EPSC’s¶
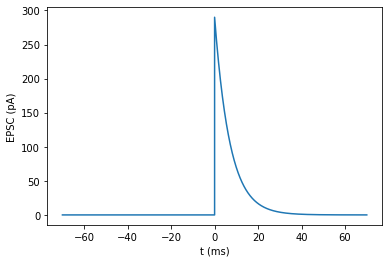
Simple step-and-exponential-decay impulse response.
Parameters estimated from fig. 5.14 in Dayan & Abott.
tau_EPSC = 7
height_EPSC = 290
T_support = 10*tau_EPSC
N_support = round(T_support/dt)
t_support = np.linspace(0, T_support, N_support)
EPSC = np.concatenate([
np.zeros(N_support),
np.exp(-t_support/tau_EPSC) * height_EPSC
])
plot_signal(EPSC, 'EPSC (pA)', t=np.concatenate([-t_support[::-1], t_support]));
I = np.convolve(all_spikes, EPSC, mode='same')
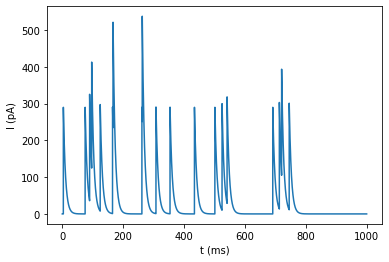
plot_signal(I, 'I (pA)');
Apply input to neuron¶
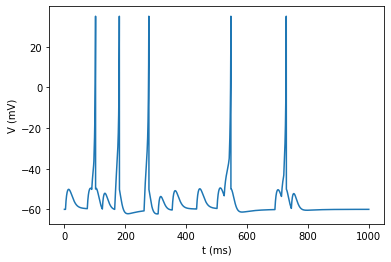
v = izh_neuron(I)
plot_signal(v, 'V (mV)');
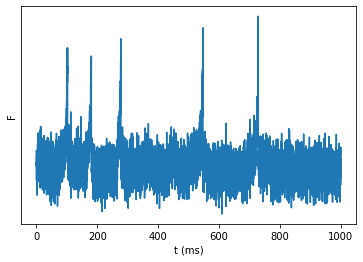
Add voltage imaging noise¶
Gaussian noise, with \(\sigma\) from:
\(\text{SNR} = \frac{ΔV_{spike}}{\sigma_{noise}}\)
(see ‘Fidelity’ section from VI lit review).
SNR = 10
spike_height = v_peak - v_r
s_noise = spike_height / SNR
noise = np.random.randn(N)*s_noise
fig, ax = plot_signal(v+noise, 'F')
ax.set_yticks([]);