2020-11-30 • Sparse spike trains
Contents
2020-11-30 • Sparse spike trains¶
Going further on /notebooks/2020-11-30__speedup, here we actually try to replace full “0/1” spike trains spike index arrays, in the entire codebase.
Imports & time grid¶
from voltage_to_wiring_sim.support.notebook_init import *
Preloading:
- numpy … (0.10 s)
- matplotlib.pyplot … (0.22 s)
- numba … (0.30 s)
Importing from submodules (compiling numba functions) … ✔
Imported `np`, `mpl`, `plt`
Imported codebase (`voltage_to_wiring_sim`) as `v`
Imported `*` from `v.support.units`
Setup autoreload
v.print_reproducibility_info()
This cell was last run by tfiers on yoga
on Tue 29 Dec 2020, at 21:23 (UTC+0100).
Last git commit (Tue 29 Dec 2020, 21:19).
Uncommited changes to:
A notebooks/2020-11-11__unitlib.ipynb
tg = v.TimeGrid(
duration = 10 * minute,
timestep = 0.1 * ms
);
Generate VI signal¶
Spike trains¶
N_in = 30
p_connected = 0.5
N_connected = round(N_in * p_connected)
N_unconnected = N_in - N_connected
15
f_spike = 20 * Hz;
gen_st = v.generate_Poisson_spikes
v.fix_rng_seed()
%%time
spike_trains_connected = [gen_st(f_spike, tg.duration) for _ in range(N_connected)]
spike_trains_unconnected = [gen_st(f_spike, tg.duration) for _ in range(N_unconnected)];
Wall time: 15 ms
This was 1.77 seconds when generating full 0/1 signals. I.e. we got 10x speedup by switching to drawing ISI’s from an exponential and only saving spike times.
all_spike_trains = spike_trains_connected + spike_trains_unconnected;
time_slice = 1 * minute + np.array([0, 1]) * second
slice_indices = np.round(time_slice / tg.timestep).astype(int)
i_slice = slice(*slice_indices)
t_slice = tg.time[i_slice];
New eventplot function:
def plot_spike_train_excerpt(spike_train):
return v.spike_trains.plot(spike_train, time_slice)
plot_spike_train_excerpt(all_spike_trains[0]);
all_incoming_spikes = np.concatenate(spike_trains_connected)
plot_spike_train_excerpt(all_incoming_spikes);
Synaptic conductance¶
Δg_syn = 0.8 * nS
τ_syn = 7 * ms;
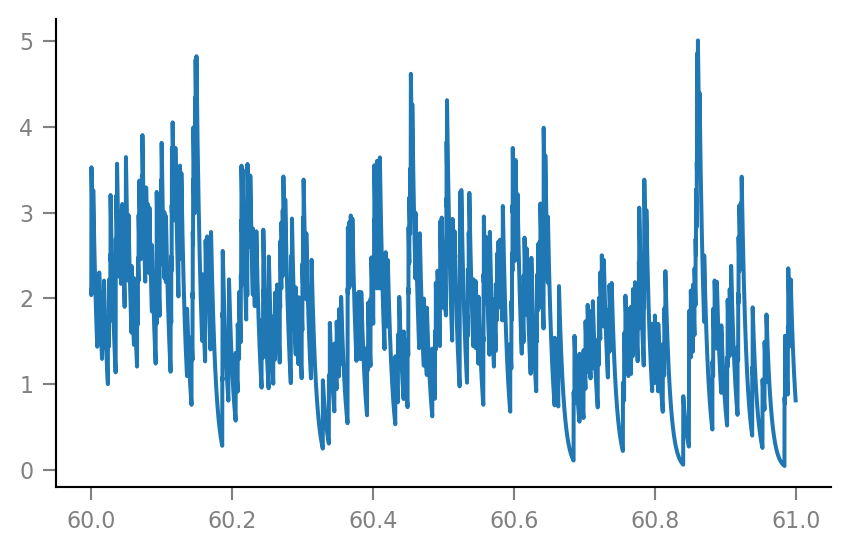
g_syn = v.calc_synaptic_conductance(tg, all_incoming_spikes, Δg_syn, τ_syn)
plt.plot(t_slice, g_syn[i_slice] / nS);
Membrane voltage¶
params = v.params.cortical_RS
v.pprint(params)
IzhikevichParams
----------------
C = 1E-10
k = 7E-07
v_r = -0.06
v_t = -0.04
v_peak = 0.035
a = 30
b = -2E-09
c = -0.05
d = 1E-10
v_syn = 0
%%time
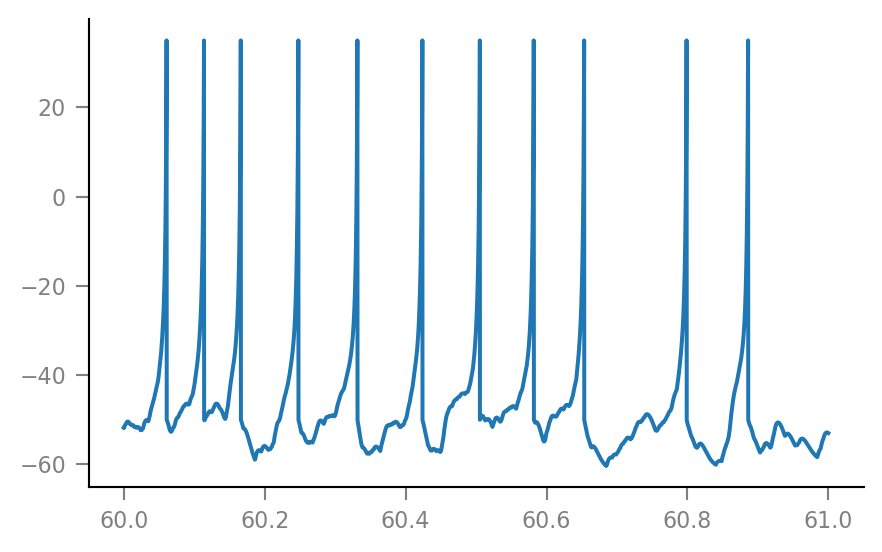
sim = v.simulate_izh_neuron(tg, params, g_syn)
Wall time: 536 ms
plt.plot(t_slice / second, sim.V_m[i_slice] / mV);
Imaging model¶
Vm_noisy = v.add_VI_noise(sim.V_m, params)
plt.plot(t_slice / second, Vm_noisy[i_slice] / mV);
Signal(data=array([-0.02985, -0.05727, -0.07003, ..., -0.04183, -0.04099, -0.05304]), timestep=0.0001)
Test connection¶
..of one spike train to imaged neuron.
spike_train = all_spike_trains[0];
%%prun -D temp.profile
v.fix_rng_seed()
test_data, test_summary = v.test_connection(
spike_train,
Vm_noisy,
window_duration=100 * ms,
num_shuffles=1000,
)
*** Profile stats marshalled to file 'temp.profile'.
v.pprint(test_data)
ConnectionTestData
------------------
shuffled_spike_trains = [array([0.0850..., 599.9, 600]), array([0.0097..., 599.8, 600]),
array([0.0090..., 599.9, 600]),
array([0.0466..., 599.9, 600]),
array([0.0309..., 599.9, 600]),
array([0.0360..., 599.9, 600]), ...]
original_STA_height = 0.0003249
shuffled_STA_heights = array([8.795E...5, 8.026E-05])
shuffled_STA_height_mean = 9.089E-05
test_data.original_STA_height / mV
0.3249126618965498
test_data.shuffled_STA_height_mean / mV
0.09089172066893161
(No easy way to make numpy numbers (“array scalars”) print nicely, i.e. with less precision; besides casting to float()).
v.pprint(test_summary)
ConnectionTestSummary
---------------------
p_value = 0.001
p_value_type = '<'
relative_STA_height = 3.575
v.test_connection with 1000 shuffles took 5.2 à 5.3 seconds (with already done jit compilation).
Most time (4.2 s) was spent in _calc_STA, i.e. an already jit-compiled function.
(after that, 0.6 and 0.3 seconds were spent in np.round and np.permutation, respectively).
Can we speedup _calc_STA even more by parallelizing the loop? The loop iterations (cut out window and add to accumulator) are independent after all.
Wait. why is it suddenly 6.3 seconds for just 100 iterations above?
-ah, maybe parallel=False is not the same as omitting parallel kwarg. Also there was the file based cache. Without those two, I get reproducible result: again 5.3 seconds for 1000 iterations, without parallel/prange.
Ok, now with parallel/prange:
%%prun -D temp.profile
v.fix_rng_seed()
test_data, test_summary = v.test_connection(
spike_train,
Vm_noisy,
window_duration=100 * ms,
num_shuffles=1000,
)
*** Profile stats marshalled to file 'temp.profile'.
We get 3.4 seconds now. A definite improvement, but not massive.
1.77 seconds in _calc_STA, but suddenly 1.14 seconds in np.round, weirdly (np.permutation is still 0.23 s). [reproduced twice].
But looking just at _calc_STA, we do get a 2x improvement.
v.pprint(test_summary)
ConnectionTestSummary
---------------------
p_value = 0.001
p_value_type = '<'
relative_STA_height = 3.575
v.STA._calc_STA.jit_compiled_function.parallel_diagnostics()
================================================================================
Parallel Accelerator Optimizing: Function _calc_STA, c:\gdrive\phd\voltage-to-
wiring-sim\codebase\voltage_to_wiring_sim\STA.py (27)
================================================================================
Parallel loop listing for Function _calc_STA, c:\gdrive\phd\voltage-to-wiring-sim\codebase\voltage_to_wiring_sim\STA.py (27)
-------------------------------------------------|loop #ID
@compile_to_machine_code(parallel=True) |
def _calc_STA( |
VI_signal: np.ndarray, |
spike_indices: np.ndarray, |
window_length: int, |
) -> np.ndarray: |
num_windows = len(spike_indices) |
STA = np.zeros(window_length)----------------| #0
for i in prange(num_windows):----------------| #2
start_ix = spike_indices[i] |
end_ix = start_ix + window_length |
if end_ix < len(VI_signal): |
STA += VI_signal[start_ix:end_ix] |
else: |
num_windows -= 1 |
return STA / num_windows---------------------| #1
------------------------------ After Optimisation ------------------------------
Parallel structure is already optimal.
--------------------------------------------------------------------------------
--------------------------------------------------------------------------------
Repro info¶
v.print_reproducibility_info(verbose=True)
This cell was last run by tfiers on yoga
on Tue 29 Dec 2020, at 21:23 (UTC+0100).
Last git commit (Tue 29 Dec 2020, 21:19).
Uncommited changes to:
A notebooks/2020-11-11__unitlib.ipynb
?? notebooks/temp.profile
Platform:
Windows-10
CPython 3.8.3 (C:\conda\python.exe)
Intel(R) Core(TM) i7-10510U CPU @ 1.80GHz
Dependencies of voltage_to_wiring_sim and their installed versions:
numpy 1.19.2
matplotlib 3.3.2
numba 0.51.2
seaborn 0.10.1
scipy 1.5.2
scikit-learn 0.23.2
preload 2.1
py-cpuinfo 7.0.0
nptyping 1.3.0