2021-11-11 • Vary both inhibitory strength and proportion
Contents
2021-11-11 • Vary both inhibitory strength and proportion¶
Prelude¶
from voltage_to_wiring_sim.notebook_init import *
Preloading: numpy, numba, matplotlib.pyplot, seaborn.
Importing from submodules … ✔
Imported `np`, `mpl`, `plt`, `sns`, `pd`
Imported codebase (`voltage_to_wiring_sim`) as `v`
Imported `*` from `v.support.units`
Setup autoreload
v.print_reproducibility_info()
This cell was last run by lpxtf3 on DUIP74576 on Thu 18 Nov 2021, at 17:18 (UTC+0000).
Last git commit (Thu 18 Nov 2021, 17:05).
Uncommited changes to 2 files.
Calculate¶
from voltage_to_wiring_sim.experiments.N_to_1_IE import Params, simulate_and_test_connections, eval_performance
base_params = Params()
v.pprint(base_params)
Params
------
sim_duration = 600
timestep = 0.0001
τ_syn = 0.007
neuron_params = {'C': 1e-10, 'a': 30.0, 'b': -2e-09, 'c': -0.05, ...}
imaging_spike_SNR = 20
window_duration = 0.1
num_spike_trains = 30
p_inhibitory = 0.2
p_connected = 0.7
spike_rate = 20
Δg_syn_exc = 4E-10
Δg_syn_inh = 1.6E-09
v_syn_exc = 0
v_syn_inh = -0.065
rng_seed = 0
@v.cache_to_disk(directory="2021-11-11__vary_both")
def sim_test_eval(params: Params):
sim_data, _, test_summaries = simulate_and_test_connections(params)
evalu_p_0_05, _, AUC, AUC_exc, AUC_inh = eval_performance(sim_data, test_summaries)
output_spike_rate = sim_data.izh_output.spike_times.size / params.sim_duration
return (output_spike_rate,
evalu_p_0_05.TPR_exc, evalu_p_0_05.TPR_inh, evalu_p_0_05.FPR,
AUC, AUC_exc, AUC_inh)
from dataclasses import replace
from itertools import product
dgsyn_IE_ratios = np.array([8, 4, 3, 2, 1, 0.5])
def get_dgsyn_vals(ratio, dgsyn_exc = 0.4 * nS):
return dict(Δg_syn_exc=dgsyn_exc,
Δg_syn_inh=dgsyn_exc * ratio)
from voltage_to_wiring_sim.experiments.N_to_1_IE import round_stochastically, fix_rng_seed
def get_N_vals(N_IE_ratio, seed, num_exc=24):
fix_rng_seed(seed)
num_inh = round_stochastically(N_IE_ratio * num_exc)
N = num_exc + num_inh
return dict(num_spike_trains=N,
p_inhibitory=num_inh/N)
N_IE_ratios = 1 / dgsyn_IE_ratios
array([0.125, 0.25, 0.3333, 0.5, 1, 2])
seeds = [0];
seeds = [0, 1, 2];
seeds = [0, 1, 2, 3, 4, 5];
# seeds = np.arange(20);
f = sim_test_eval
args = [replace(base_params,
**get_dgsyn_vals(dgsyn_IE_ratio),
**get_N_vals(N_IE_ratio),
rng_seed=seed,
)
for (dgsyn_IE_ratio, N_IE_ratio, seed) in product(dgsyn_IE_ratios, N_IE_ratios, seeds)];
%%time
results = v.run_in_parallel(f, args);
Wall time: 13min 38s
(eg: 6 minutes for 4x5 params and 3 seeds, using 8 cores)
Or: 19min 30s for 6x6 params and 3 seeds (but diagonal was already calculated).
M = np.reshape(results, (len(dgsyn_IE_ratios), len(N_IE_ratios), len(seeds), -1))
(output_spike_rate,
TPR_exc, TPR_inh, FPR,
AUC, AUC_exc, AUC_inh) = (M[:,:,:,i] for i in range(M.shape[-1]))
Plot¶
def matplot(data, title: str, ax, cmap="plasma", vmin=None, vmax=None):
data = np.fliplr(data.T)
im = ax.imshow(data, origin="upper", cmap=cmap, vmin=vmin, vmax=vmax)
ax.set_xticks(range(len(dgsyn_IE_ratios)))
ax.set_xticklabels(f"{d:g}" for d in reversed(dgsyn_IE_ratios))
ax.set_xlabel("Inhibitory connection" "\n" "strength $\; Δg_{inh} \; / \; Δg_{exc}$")
ax.set_yticks(range(len(N_IE_ratios)))
ax.set_yticklabels(f"{p:.2g}" for p in N_IE_ratios)
ax.set_ylabel("Inhibitory inputs $\; N_{inh} \; / \; N_{exc}$")
ax.grid(False)
ax.set_title(title, pad=10, size="medium")
for row, col in list(product(*(range(d) for d in data.shape))):
val = data[row, col]
text = f"{val:.2g}"
size = "small" if len(text) <= 5 else "x-small"
color = "0.3" if im.norm(val) > 0.5 else "0.7"
t = ax.text(col, row, text, ha="center", va="center", color=color, size=size)
matplot_AUC = partial(matplot, vmin=0.5, vmax=1, cmap="cividis")
fig, axes = plt.subplots(ncols=3, **v.figsize(width=1000, aspect=1.8))
matplot(np.mean(output_spike_rate, axis=-1), "Output spike rate (Hz)", axes[0], cmap="plasma")
matplot_AUC(np.mean(AUC_exc, axis=-1), "Detection ability for" "\n" "excitatory connections (AUC)", axes[1])
matplot_AUC(np.mean(AUC_inh, axis=-1), "Detection ability for" "\n" "inhibitory connections (AUC)", axes[2])
axes[1].set_ylabel(None)
axes[2].set_ylabel(None);
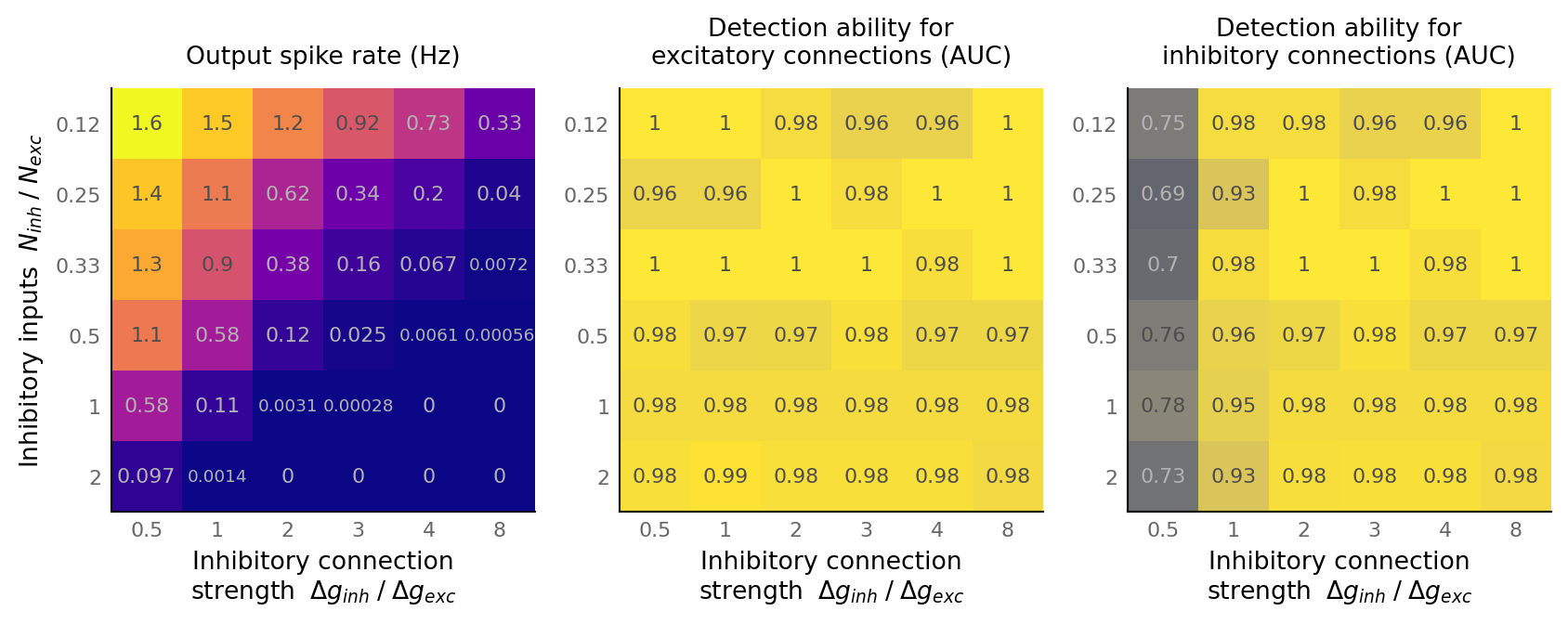
fig, axes = plt.subplots(ncols=3, **v.figsize(width=1000, aspect=1.8))
def cov(data):
return np.std(data, axis=-1) / np.mean(data, axis=-1)
matplot(cov(output_spike_rate), "", axes[0], vmin=0, vmax=0.3, cmap='viridis')
matplot(cov(AUC_exc), "", axes[1], vmin=0, vmax=0.3, cmap='viridis')
matplot(cov(AUC_inh), "", axes[2], vmin=0, vmax=0.3, cmap='viridis')
axes[1].set_ylabel(None)
axes[2].set_ylabel(None)
fig.suptitle("Coefficients of variation", y=0.8);
C:\Users\lpxtf3\AppData\Local\Temp/ipykernel_120568/1577590883.py:4: RuntimeWarning: invalid value encountered in true_divide
return np.std(data, axis=-1) / np.mean(data, axis=-1)
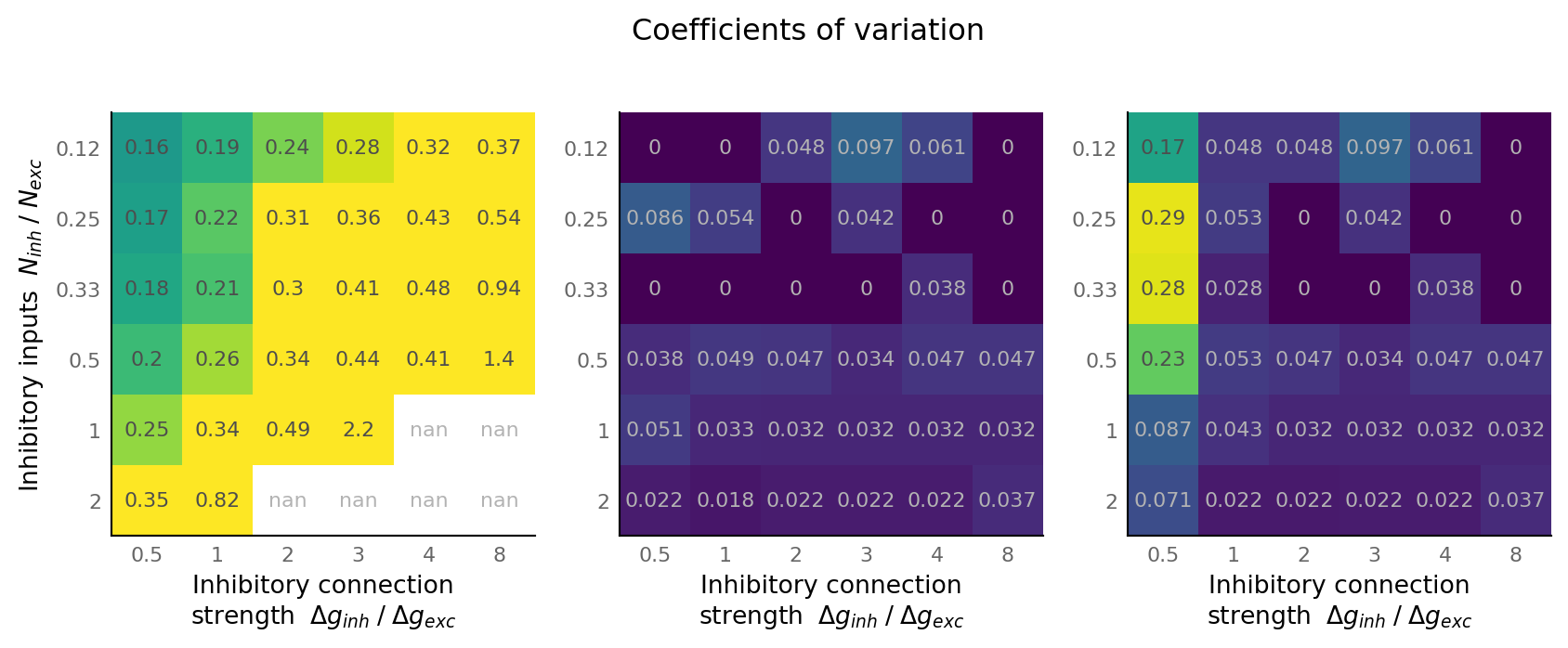
fig, axes = plt.subplots(ncols=3, **v.figsize(width=1000, aspect=1.8))
spike_range = dict(vmin=output_spike_rate.min(), vmax=output_spike_rate.max())
matplot(np.min(output_spike_rate, axis=-1), "", axes[0], cmap="plasma", **spike_range)
matplot_AUC(np.min(AUC_exc, axis=-1), "", axes[1])
matplot_AUC(np.min(AUC_inh, axis=-1), "", axes[2])
axes[1].set_ylabel(None)
axes[2].set_ylabel(None)
fig.suptitle("Minimum values", y=0.77)
fig, axes = plt.subplots(ncols=3, **v.figsize(width=1000, aspect=1.8))
matplot(np.max(output_spike_rate, axis=-1), "", axes[0], cmap="plasma", **spike_range)
matplot_AUC(np.max(AUC_exc, axis=-1), "", axes[1])
matplot_AUC(np.max(AUC_inh, axis=-1), "", axes[2])
axes[1].set_ylabel(None)
axes[2].set_ylabel(None)
fig.suptitle("Maximum values", y=0.77);
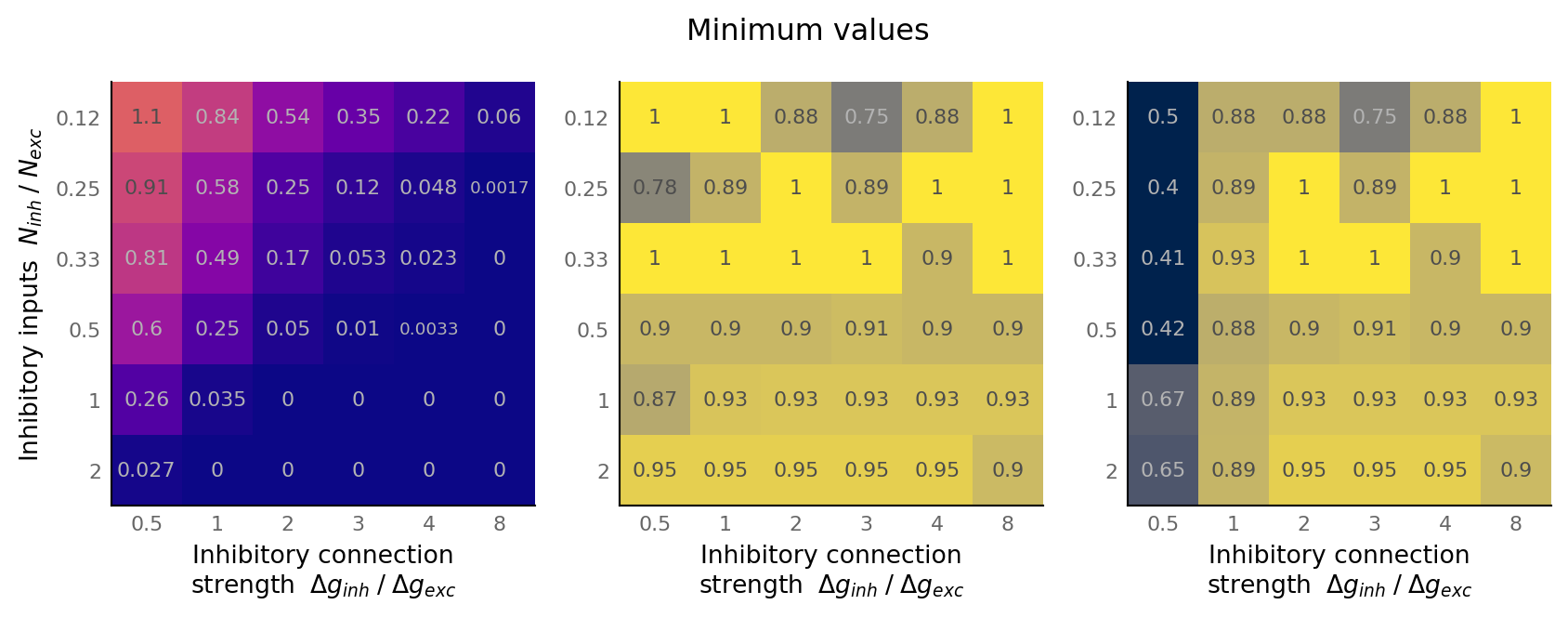
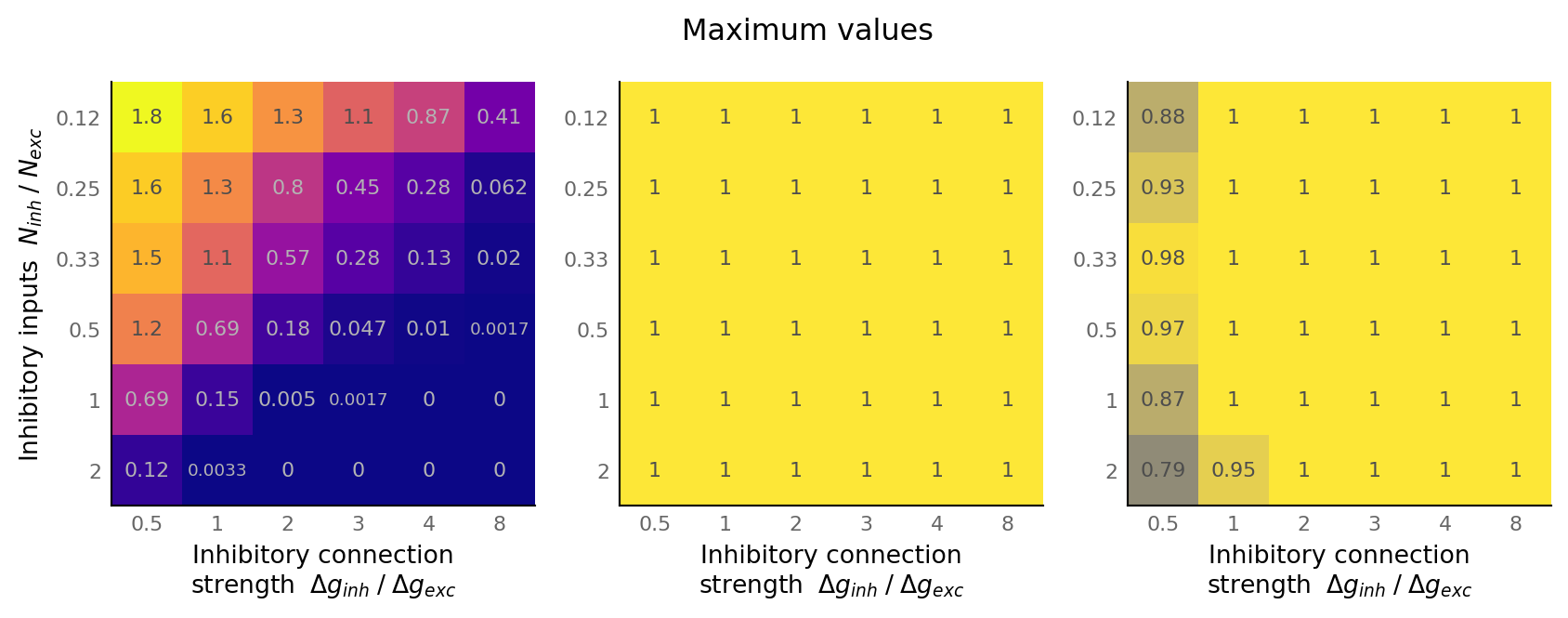
Diagonal only¶
dgsyn_IE_ratios_diag = np.array([8, 6, 4, 3, 2, 1, 0.5]);
seeds_diag = [0];
seeds_diag = [0, 1, 2, 3, 4, 5];
seeds_diag = np.arange(20);
args_diag = [replace(base_params,
**get_dgsyn_vals(dgsyn_IE_ratio),
**get_N_vals(1 / dgsyn_IE_ratio, seed),
rng_seed=seed,
)
for (dgsyn_IE_ratio, seed) in product(dgsyn_IE_ratios_diag, seeds_diag)];
%%time
results_diag = v.run_in_parallel(f, args_diag);
Wall time: 221 ms
M_diag = np.reshape(results_diag, (len(dgsyn_IE_ratios_diag), len(seeds_diag), -1))
(output_spike_rate,
TPR_exc, TPR_inh, FPR,
AUC, AUC_exc, AUC_inh) = (M_diag[:,:,i] for i in range(M_diag.shape[-1]))
def plot_dots_and_mean(data, ax, x, c="0.2", label=None, ms=6, lw=5):
ax.plot(x, data, "o", alpha=0.3, c=c, ms=ms)
ax.plot(x, np.mean(data, axis=-1), "-", c=c, label=label, lw=lw)
smaller = dict(ms=4, lw=3);
def make_inv_ratio_ax():
fig, ax = plt.subplots()
ax.set_xscale('log')
ax.set_xticks(dgsyn_IE_ratios_diag)
ax.set_xticklabels(f"{r:g}" for r in dgsyn_IE_ratios_diag)
ax.set_xlabel("Inhibitory connection strength $\; Δg_{inh} \; / \; Δg_{exc}$")
# https://matplotlib.org/stable/gallery/subplots_axes_and_figures/secondary_axis.html
def one_over(x):
x = np.array(x).astype(float)
near_zero = np.isclose(x, 0)
x[near_zero] = np.inf
x[~near_zero] = 1 / x[~near_zero]
return x
sax = ax.secondary_xaxis("top", functions=(one_over, one_over))
N_IE_ratios_diag = 1 / dgsyn_IE_ratios_diag
sax.set_xticks(N_IE_ratios_diag)
sax.set_xticks([], minor=True)
sax.set_xticklabels(f"{r:.3g}" for r in N_IE_ratios_diag)
sax.set_xlabel("Number of inhibitory inputs $\; N_{inh} \; / \; N_{exc}$", labelpad=8)
return fig, ax
plot = partial(plot_dots_and_mean, x=dgsyn_IE_ratios_diag);
fig, ax = make_inv_ratio_ax()
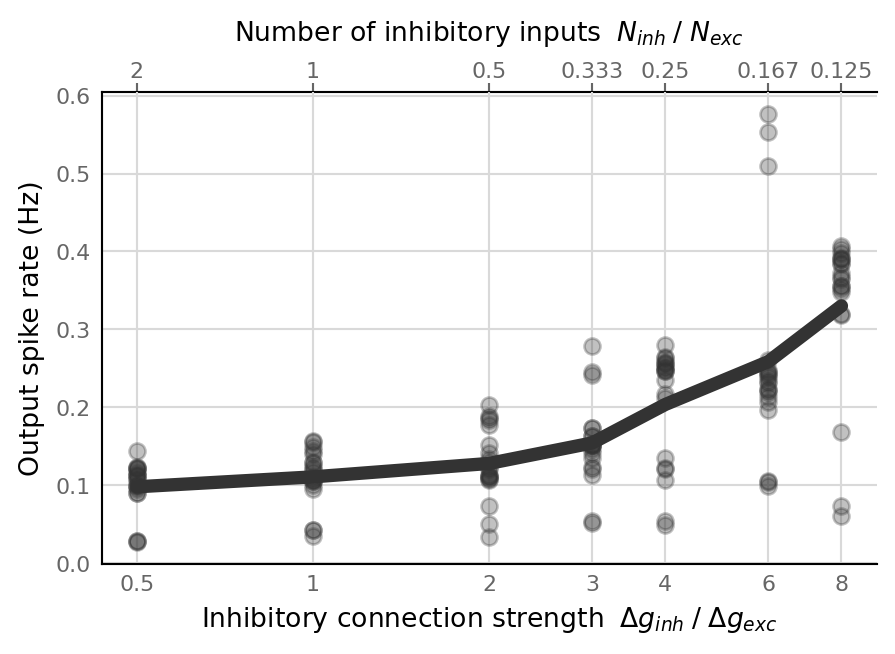
plot(output_spike_rate, ax)
ax.set_ylabel("Output spike rate (Hz)");
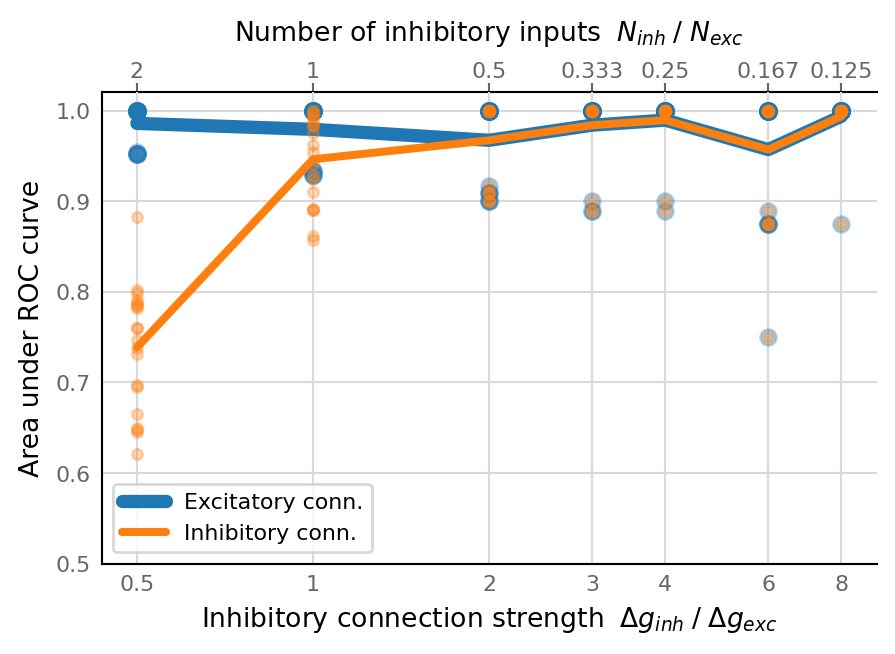
fig, ax = make_inv_ratio_ax()
plot(AUC_exc, ax, c=v.color_exc, label="Excitatory conn.")
plot(AUC_inh, ax, c=v.color_inh, label="Inhibitory conn.", **smaller)
ax.set_ylim(0.5, 1.02)
ax.set_ylabel("Area under ROC curve")
ax.legend();
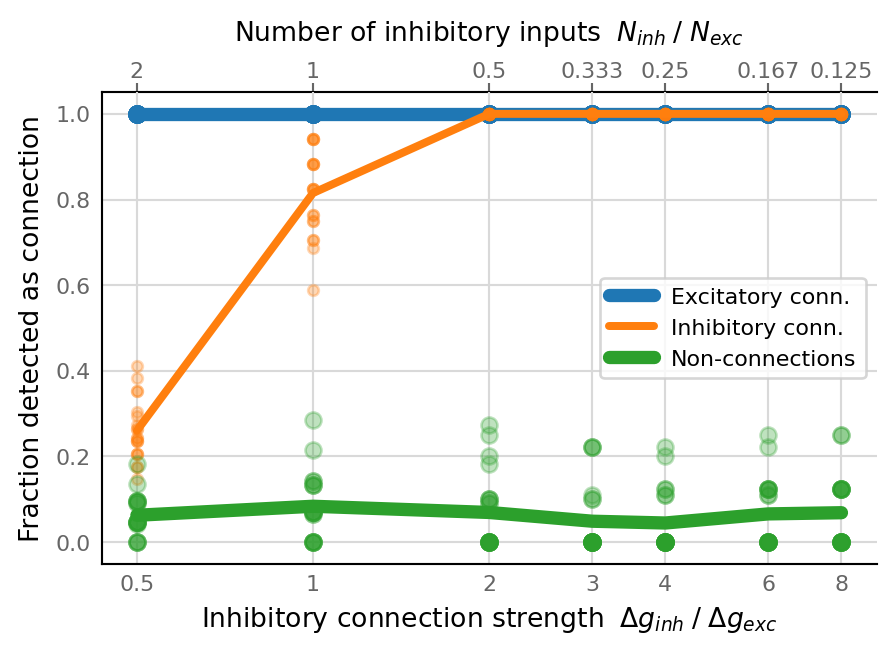
fig, ax = make_inv_ratio_ax()
plot(TPR_exc, ax, c=v.color_exc, label="Excitatory conn.")
plot(TPR_inh, ax, c=v.color_inh, label="Inhibitory conn.", **smaller)
plot(FPR, ax, c=v.color_unconn, label="Non-connections")
ax.set_ylabel("Fraction detected as connection")
ax.legend();
Inspect signals¶
sim_and_test = v.cache_to_disk(simulate_and_test_connections);
dgsyn_ratio = 8
d_8, td_8, ts_8 = sim_and_test(replace(base_params, **get_dgsyn_vals(dgsyn_ratio), **get_N_vals(1 / dgsyn_ratio)));
dgsyn_ratio = 1
d_1, td_1, ts_1 = sim_and_test(replace(base_params, **get_dgsyn_vals(dgsyn_ratio), **get_N_vals(1 / dgsyn_ratio)));
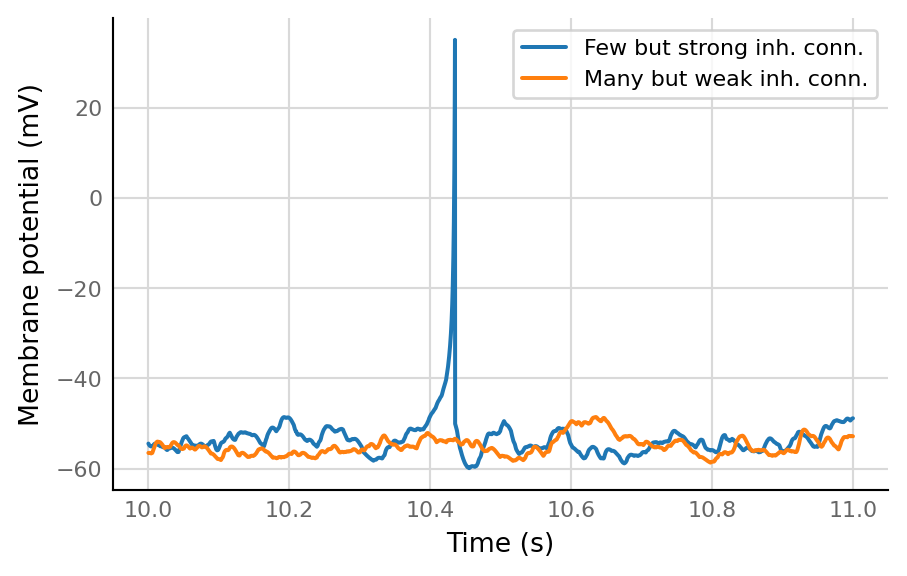
def plot_slice(sig, ax, **kwargs):
v.plot_signal(sig.slice(10*second, 1*second), ax, **kwargs)
fig, ax = plt.subplots()
plot_slice(d_8.izh_output.V_m / mV, ax, label="Few but strong inh. conn.")
plot_slice(d_1.izh_output.V_m / mV, ax, label="Many but weak inh. conn.")
ax.legend()
ax.set(xlabel="Time (s)", ylabel="Membrane potential (mV)");
Total conductance¶
from voltage_to_wiring_sim.experiments.N_to_1_IE import indices_where
d_8.num_exc_conn, d_8.num_inh_conn
(17, 2)
d_1.num_exc_conn, d_1.num_inh_conn
(17, 17)
g_inh_8 = sum(d_8.g_syns[i] for i in indices_where(d_8.is_inhibitory[d_8.is_connected]));
g_inh_1 = sum(d_1.g_syns[i] for i in indices_where(d_1.is_inhibitory[d_1.is_connected]));
fig, ax = plt.subplots()
plot_slice(g_inh_8 / nS, ax)
plot_slice(g_inh_1 / nS, ax)
ax.axhline(np.mean(g_inh_8) / nS, c="C0")
ax.axhline(np.mean(g_inh_1) / nS, c="C1")
ax.set(xlabel="Time (s)", ylabel="Inh. synaptic conductance (nS)");
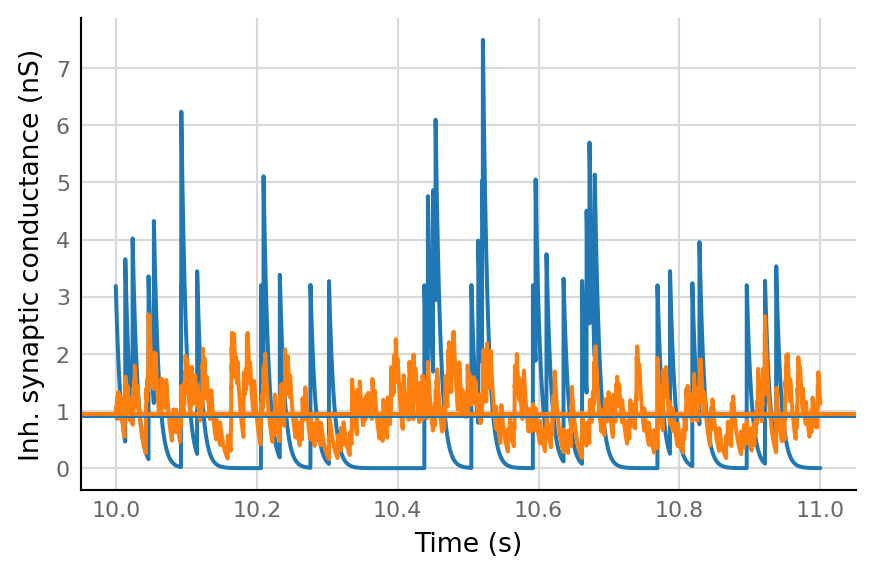
(Medians differ, but means are the same).
g_exc_8 = sum(d_8.g_syns[i] for i in indices_where(d_8.is_excitatory[d_8.is_connected]));
g_exc_1 = sum(d_1.g_syns[i] for i in indices_where(d_1.is_excitatory[d_1.is_connected]));
fig, ax = plt.subplots()
plot_slice(g_exc_8 / nS, ax)
plot_slice(g_exc_1 / nS, ax)
ax.axhline(np.median(g_exc_8) / nS, c="C0")
ax.axhline(np.median(g_exc_1) / nS, c="C1");
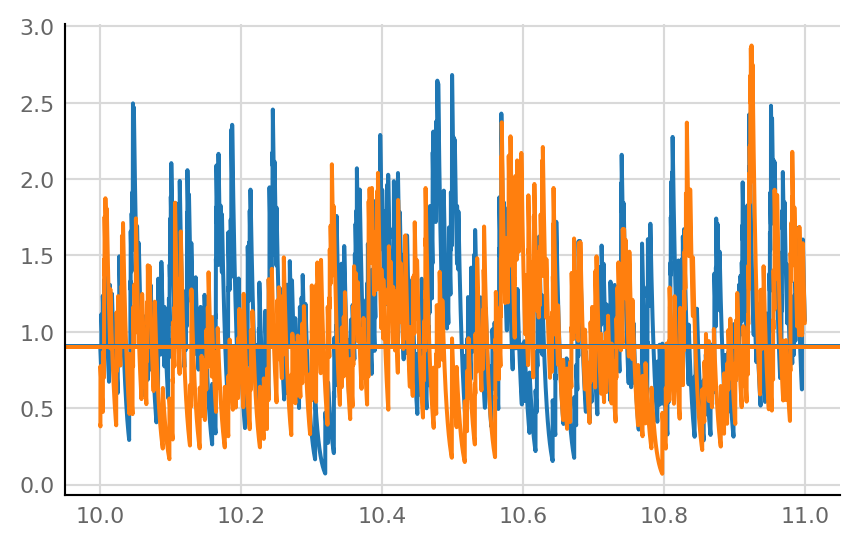
Reproducibility¶
v.print_reproducibility_info(verbose=True)
This cell was last run by lpxtf3 on DUIP74576
on Thu 18 Nov 2021, at 16:55 (UTC+0000).
Last git commit (Fri 12 Nov 2021, 00:23).
Uncommited changes to:
M ReadMe.md
M codebase/voltage_to_wiring_sim/__init__.py
M codebase/voltage_to_wiring_sim/experiments/N_to_1.py
M codebase/voltage_to_wiring_sim/support/__init__.py
AM codebase/voltage_to_wiring_sim/support/high_performance.py
M codebase/voltage_to_wiring_sim/support/misc.py
M notebooks/2021-01-02__full_network_sim_tryout.ipynb
M notebooks/2021-11-11__vary_both_inh_strength_and_proportion.ipynb
M website/thesis/references.bib
Platform:
Windows-10
CPython 3.9.6 (C:\miniforge3\python.exe)
Intel(R) Xeon(R) W-2123 CPU @ 3.60GHz
Dependencies of voltage_to_wiring_sim and their installed versions:
numpy 1.21.1
matplotlib 3.4.2
numba 0.53.1
joblib 1.0.1
seaborn 0.11.1
scipy 1.7.0
preload 2.2
nptyping 1.4.2
Full conda list:
# packages in environment at C:\miniforge3:
#
# Name Version Build Channel
argon2-cffi 20.1.0 py39hb82d6ee_2 conda-forge
async_generator 1.10 py_0 conda-forge
attrs 21.2.0 pyhd8ed1ab_0 conda-forge
backcall 0.2.0 pyh9f0ad1d_0 conda-forge
backports 1.0 py_2 conda-forge
backports.functools_lru_cache 1.6.4 pyhd8ed1ab_0 conda-forge
black 21.9b0 pyhd8ed1ab_1 conda-forge
bleach 3.3.1 pyhd8ed1ab_0 conda-forge
brotlipy 0.7.0 py39hb82d6ee_1001 conda-forge
ca-certificates 2021.10.8 h5b45459_0 conda-forge
certifi 2021.10.8 py39hcbf5309_1 conda-forge
cffi 1.14.6 py39h0878f49_0 conda-forge
chardet 4.0.0 py39hcbf5309_1 conda-forge
charset-normalizer 2.0.0 pyhd8ed1ab_0 conda-forge
click 7.1.2 pypi_0 pypi
colorama 0.4.4 pyh9f0ad1d_0 conda-forge
colorful 0.5.4 pypi_0 pypi
conda 4.10.3 py39hcbf5309_2 conda-forge
conda-package-handling 1.7.3 py39hb3671d1_0 conda-forge
cryptography 3.4.7 py39hd8d06c1_0 conda-forge
cycler 0.10.0 pypi_0 pypi
dataclasses 0.8 pyhc8e2a94_3 conda-forge
debugpy 1.4.1 py39h415ef7b_0 conda-forge
decorator 5.0.9 pyhd8ed1ab_0 conda-forge
defusedxml 0.7.1 pyhd8ed1ab_0 conda-forge
entrypoints 0.3 pyhd8ed1ab_1003 conda-forge
icu 68.2 h0e60522_0 conda-forge
idna 3.1 pyhd3deb0d_0 conda-forge
importlib-metadata 4.6.1 py39hcbf5309_0 conda-forge
ipykernel 6.0.3 py39h832f523_0 conda-forge
ipython 7.25.0 py39h832f523_1 conda-forge
ipython_genutils 0.2.0 py_1 conda-forge
jedi 0.18.0 py39hcbf5309_2 conda-forge
jinja2 3.0.1 pyhd8ed1ab_0 conda-forge
joblib 1.0.1 pypi_0 pypi
jpeg 9d h8ffe710_0 conda-forge
jsonschema 3.2.0 pyhd8ed1ab_3 conda-forge
jupyter_client 6.1.12 pyhd8ed1ab_0 conda-forge
jupyter_contrib_core 0.3.3 py_2 conda-forge
jupyter_contrib_nbextensions 0.5.1 pyhd8ed1ab_2 conda-forge
jupyter_core 4.7.1 py39hcbf5309_0 conda-forge
jupyter_highlight_selected_word 0.2.0 py39hcbf5309_1002 conda-forge
jupyter_latex_envs 1.4.6 pyhd8ed1ab_1002 conda-forge
jupyter_nbextensions_configurator 0.4.1 py39hcbf5309_2 conda-forge
jupyterlab_pygments 0.1.2 pyh9f0ad1d_0 conda-forge
kiwisolver 1.3.1 pypi_0 pypi
libclang 11.1.0 default_h5c34c98_1 conda-forge
libiconv 1.16 he774522_0 conda-forge
libpng 1.6.37 h1d00b33_2 conda-forge
libsodium 1.0.18 h8d14728_1 conda-forge
libxml2 2.9.12 hf5bbc77_0 conda-forge
libxslt 1.1.33 h65864e5_2 conda-forge
libzlib 1.2.11 h8ffe710_1013 conda-forge
llvmlite 0.36.0 pypi_0 pypi
lxml 4.6.3 py39h4fd7cdf_0 conda-forge
markupsafe 2.0.1 py39hb82d6ee_0 conda-forge
matplotlib 3.4.2 pypi_0 pypi
matplotlib-inline 0.1.2 pyhd8ed1ab_2 conda-forge
menuinst 1.4.17 py39hcbf5309_1 conda-forge
miniforge_console_shortcut 2.0 h57928b3_0 conda-forge
mistune 0.8.4 py39hb82d6ee_1004 conda-forge
mypy_extensions 0.4.3 py39hcbf5309_4 conda-forge
nbclient 0.5.3 pyhd8ed1ab_0 conda-forge
nbconvert 6.1.0 py39hcbf5309_0 conda-forge
nbformat 5.1.3 pyhd8ed1ab_0 conda-forge
nest-asyncio 1.5.1 pyhd8ed1ab_0 conda-forge
notebook 6.4.0 pyha770c72_0 conda-forge
nptyping 1.4.2 pypi_0 pypi
numba 0.53.1 pypi_0 pypi
numpy 1.21.1 pypi_0 pypi
openssl 1.1.1l h8ffe710_0 conda-forge
packaging 21.0 pyhd8ed1ab_0 conda-forge
pandas 1.3.1 pypi_0 pypi
pandoc 2.14.1 h8ffe710_0 conda-forge
pandocfilters 1.4.2 py_1 conda-forge
parso 0.8.2 pyhd8ed1ab_0 conda-forge
pathspec 0.9.0 pyhd8ed1ab_0 conda-forge
pickleshare 0.7.5 py_1003 conda-forge
pillow 8.3.1 pypi_0 pypi
pip 21.2.1 pyhd8ed1ab_0 conda-forge
platformdirs 2.3.0 pyhd8ed1ab_0 conda-forge
preload 2.2 pypi_0 pypi
prettyprinter 0.18.0 pypi_0 pypi
prometheus_client 0.11.0 pyhd8ed1ab_0 conda-forge
prompt-toolkit 3.0.19 pyha770c72_0 conda-forge
pycosat 0.6.3 py39hb82d6ee_1006 conda-forge
pycparser 2.20 pyh9f0ad1d_2 conda-forge
pygments 2.9.0 pyhd8ed1ab_0 conda-forge
pympler 0.9 pypi_0 pypi
pyopenssl 20.0.1 pyhd8ed1ab_0 conda-forge
pyparsing 2.4.7 pyh9f0ad1d_0 conda-forge
pyqt 5.12.3 py39hcbf5309_7 conda-forge
pyqt-impl 5.12.3 py39h415ef7b_7 conda-forge
pyqt5-sip 4.19.18 py39h415ef7b_7 conda-forge
pyqtchart 5.12 py39h415ef7b_7 conda-forge
pyqtwebengine 5.12.1 py39h415ef7b_7 conda-forge
pyrsistent 0.17.3 py39hb82d6ee_2 conda-forge
pysocks 1.7.1 py39hcbf5309_3 conda-forge
python 3.9.6 h7840368_1_cpython conda-forge
python-dateutil 2.8.2 pyhd8ed1ab_0 conda-forge
python_abi 3.9 2_cp39 conda-forge
pytz 2021.1 pypi_0 pypi
pywin32 300 py39hb82d6ee_0 conda-forge
pywinpty 1.1.3 py39h99910a6_0 conda-forge
pyyaml 6.0 py39hb82d6ee_0 conda-forge
pyzmq 22.1.0 py39he46f08e_0 conda-forge
qt 5.12.9 h5909a2a_4 conda-forge
regex 2021.10.23 py39hb82d6ee_1 conda-forge
requests 2.26.0 pyhd8ed1ab_0 conda-forge
ruamel_yaml 0.15.80 py39hb82d6ee_1004 conda-forge
scipy 1.7.0 pypi_0 pypi
seaborn 0.11.1 pypi_0 pypi
send2trash 1.7.1 pyhd8ed1ab_0 conda-forge
setuptools 49.6.0 py39hcbf5309_3 conda-forge
six 1.16.0 pyh6c4a22f_0 conda-forge
sqlite 3.36.0 h8ffe710_0 conda-forge
terminado 0.10.1 py39hcbf5309_0 conda-forge
testpath 0.5.0 pyhd8ed1ab_0 conda-forge
tomli 1.2.2 pyhd8ed1ab_0 conda-forge
tornado 6.1 py39hb82d6ee_1 conda-forge
tqdm 4.61.2 pyhd8ed1ab_1 conda-forge
traitlets 5.0.5 py_0 conda-forge
typed-ast 1.4.3 py39hb82d6ee_1 conda-forge
typing_extensions 3.10.0.2 pyha770c72_0 conda-forge
typish 1.9.2 pypi_0 pypi
tzdata 2021a he74cb21_1 conda-forge
ucrt 10.0.20348.0 h57928b3_0 conda-forge
urllib3 1.26.6 pyhd8ed1ab_0 conda-forge
vc 14.2 hb210afc_5 conda-forge
voltage-to-wiring-sim 0.1 dev_0 <develop>
vs2015_runtime 14.29.30037 h902a5da_5 conda-forge
wcwidth 0.2.5 pyh9f0ad1d_2 conda-forge
webencodings 0.5.1 py_1 conda-forge
wheel 0.36.2 pyhd3deb0d_0 conda-forge
win_inet_pton 1.1.0 py39hcbf5309_2 conda-forge
wincertstore 0.2 py39hcbf5309_1006 conda-forge
winpty 0.4.3 4 conda-forge
winshell 0.6 pypi_0 pypi
yaml 0.2.5 he774522_0 conda-forge
zeromq 4.3.4 h0e60522_0 conda-forge
zipp 3.5.0 pyhd8ed1ab_0 conda-forge
zlib 1.2.11 h8ffe710_1013 conda-forge