2023-02-07 • AdEx Nto1
Contents
2023-02-07 • AdEx Nto1¶
(Based on 2023-01-19__[input], which is a distillation of 2022-10-24 • N-to-1 with lognormal inputs).
cd("/root/phd/pkg/SpikeWorks")
run(`git switch metdeklak`);
Your branch is up to date with 'origin/metdeklak'.
Already on 'metdeklak'
Imports¶
#
@showtime using Revise
using Revise: 0.599281 seconds (603.53 k allocations: 36.640 MiB, 3.70% gc time, 1.04% compilation time)
@showtime using MyToolbox
@showtime using SpikeWorks
@showtime using Sciplotlib
@showtime using VoltoMapSim
WARNING: using MyToolbox.@withfb in module Main conflicts with an existing identifier.
using MyToolbox: 1.574102 seconds (1.63 M allocations: 104.069 MiB, 2.92% gc time, 0.22% compilation time)
using SpikeWorks: 0.875073 seconds (1.28 M allocations: 78.734 MiB, 5.64% gc time)
using Sciplotlib: 9.385032 seconds (7.10 M allocations: 455.172 MiB, 2.42% gc time, 1.97% compilation time: 100% of which was recompilation)
using VoltoMapSim: 4.136105 seconds (4.98 M allocations: 328.876 MiB, 4.23% gc time)
AdEx equations & params¶
This is a distillation of previous notebook.
We base ourselves on
Richard Naud, Nicolas Marcille, Claudia Clopath, and Wulfram Gerstner,
‘Firing patterns in the adaptive exponential integrate-and-fire model’,
Biol Cybern, Nov. 2008, https://doi.org/10.1007/s00422-008-0264-7
..and their cortical RS (regular spiking) neuron in table 1.
Param value comparison, for cortical RS neuron¶
(Table repeted from prev notebook)
Naud 2008 AdEx |
Val |
Val |
Izh / report |
What |
|---|---|---|---|---|
\(C\) |
104 pF |
100 pF |
\(C\) |
|
\(C/g_L\) |
24 ms |
? |
? |
Time constant of voltage |
0.14 ms·mV |
\(C/k\) |
? |
||
————– |
——— |
———— |
————– |
|
\(g_L\) |
4.3 nS |
14 nS |
\(k(v_t-v_r)\) |
Slope of V̇(V) at rest |
\(E_L\) |
-65 mV |
-60 mV |
\(v_r\) |
Rest (stable fixed point) |
\(V_T\) |
-52 mV |
-50 mV |
\((v_t+v_r)/2\) |
Minimum of V̇(V) |
-49.6 mV |
-40 mV |
\(v_t\) |
Threshold (unstable fixed point) |
|
82 nS |
14 nS |
\(k(v_t-v_r)\) |
Slope of V̇(V) at threshold |
|
\(Δ_T\) |
0.8 mV |
|||
————– |
——— |
———— |
————– |
|
\(a\) |
-0.8 nS |
-2 nS |
\(b\) |
Sensitivity of adapt. current |
\(τ_w\) |
88 ms |
33 ms |
\(a^{-1}\) |
Time ct of adapt. current |
\(b\) |
65 pA |
100 pA |
\(d\) |
Adapt. current bump after spike |
\(V_r\) |
-53 mV |
-50 mV |
\(c\) |
Reset voltage after spike |
Start of code¶
@typed begin
# AdEx LIF neuron params (cortical RS)
C = 104 * pF
gₗ = 4.3 * nS
Eₗ = -65 * mV
Vₜ = -52 * mV
Δₜ = 0.8 * mV
Vₛ = 0 * mV
Vᵣ = -53 * mV
a = 0.8 * nS
b = 65 * pA
τw = 88 * ms
# Conductance-based synapses
Eₑ = 0 * mV
Eᵢ = -80 * mV
τ = 7 * ms
end;
Simulated variables and their initial values¶
x₀ = (
# AdEx variables
v = Vᵣ, # Membrane potential
w = 0 * pA, # Adaptation current
# Synaptic conductances g
gₑ = 0 * nS, # = Sum over all exc. synapses
gᵢ = 0 * nS, # = Sum over all inh. synapses
);
if \(V > 0\) mV, then
Differential equations:¶
calculate time derivatives of simulated vars
(and store them “in-place”, in Dₜ).
function f!(Dₜ, vars)
v, w, gₑ, gᵢ = vars
# Conductance-based synaptic current
Iₛ = gₑ*(v-Eₑ) + gᵢ*(v-Eᵢ)
# AdEx 2D system
Dₜ.v = (-gₗ*(v-Eₗ) + gₗ*Δₜ*exp((v-Vₜ)/Δₜ) - Iₛ - w) / C
Dₜ.w = (a*(v-Eₗ) - w) / τw
# Synaptic conductance decay
Dₜ.gₑ = -gₑ / τ
Dₜ.gᵢ = -gᵢ / τ
end;
We correct the sign of Iₛ.
From prev nb:
Positive charges flowing out of membrane: Iₘₑₘ pos.
Positive charges flowing from electrode into cell: Iₑₓₜ pos.
v is usually < Eₑ,
so (v-Eₑ) will be negative.
But it should be positive, if we want to have + Iₛ in our Dₜ.v equation.
Eh no, I’ll keep as is.
Consistency with Iₘₑₘ (but against convention, yes).
Spike discontinuity¶
has_spiked(vars) = (vars.v > Vₛ)
function on_self_spike!(vars)
vars.v = Vᵣ
vars.w += b
end;
Conductance-based AdEx neuron¶
coba_adex_neuron = NeuronModel(x₀, f!; has_spiked, on_self_spike!);
The rest (i.e. the Nto1 part, with E:I) is same
More parameters, and input spikers¶
using SpikeWorks.Units
using SpikeWorks: LogNormal
@typed begin
Δt = 0.1ms
sim_duration = 10minutes
end
600
Firing rates λ for the Poisson inputs
fr_distr = LogNormal(median = 4Hz, g = 2)
Distributions.LogNormal{Float64}(μ=1.39, σ=0.693)
@enum NeuronType exc inh
input(;
N = 100,
EIratio = 4//1,
scaling = N,
) = begin
firing_rates = rand(fr_distr, N)
input_IDs = 1:N
inputs = [
Nto1Input(ID, poisson_SpikeTrain(λ, sim_duration))
for (ID, λ) in zip(input_IDs, firing_rates)
]
# Nₑ, Nᵢ = groupsizes(EIMix(N, EIratio))
EImix = EIMix(N, EIratio)
Nₑ = EImix.Nₑ
Nᵢ = EImix.Nᵢ
neuron_type(ID) = (ID ≤ Nₑ) ? exc : inh
Δgₑ = 60nS / scaling
Δgᵢ = 60nS / scaling * EIratio
on_spike_arrival!(vars, spike) =
if neuron_type(source(spike)) == exc
vars.gₑ += Δgₑ
else
vars.gᵢ += Δgᵢ
end
return (;
firing_rates,
inputs,
on_spike_arrival!,
Nₑ,
)
end;
using SpikeWorks: Simulation, step!, run!, unpack, newsim,
get_new_spikes!, next_spike, index_of_next
new(; kw...) = begin
ip = input(; kw...)
s = newsim(coba_adex_neuron, ip.inputs, ip.on_spike_arrival!, Δt)
(sim=s, input=ip)
end;
Multi sim¶
(These Ns are same as in e.g. https://tfiers.github.io/phd/nb/2022-10-11__Nto1_output_rate__Edit_of_2022-05-02.html)
using SpikeWorks: spikerate
sim_duration/minutes
10
using Printf
print_Δt(t0) = @printf("%.2G seconds\n", time()-t0)
macro timeh(ex) :( t0=time(); $(esc(ex)); print_Δt(t0) ) end;
Ns_and_scalings = [
(5, 2.4), # => N_inh = 1
(20, 1.3),
# orig: 21.
# But: "pₑ = 0.8 does not divide N = 21 into integer parts"
# So voila
(100, 0.8),
(400, 0.6),
(1600, 0.5),
(6500, 0.5),
];
Ns = first.(Ns_and_scalings);
nbname = "2023-02-07__AdEx_Nto1"
cachekey(N) = "$(nbname)__N=$(N)__T=$(sim_duration)";
cachekey(Ns[end])
"2023-02-07__AdEx_Nto1__N=6500__T=600"
function runsim(N, scaling)
println()
(sim, inp) = new(; N, scaling)
@show N
@timeh run!(sim)
@show spikerate(sim)
return (; sim, input=inp)
end
simruns = []
for (N, f) in Ns_and_scalings
scaling = f*N
simrun = cached(runsim, (N, scaling), key=cachekey(N))
push!(simruns, simrun)
end
Loading cached output from `/root/.phdcache/runsim/2023-02-07__AdEx_Nto1__N=5__T=600.jld2` … done (5.4 s)
Loading cached output from `/root/.phdcache/runsim/2023-02-07__AdEx_Nto1__N=20__T=600.jld2` … done (0.1 s)
Loading cached output from `/root/.phdcache/runsim/2023-02-07__AdEx_Nto1__N=100__T=600.jld2` … done (0.1 s)
Loading cached output from `/root/.phdcache/runsim/2023-02-07__AdEx_Nto1__N=400__T=600.jld2` … done (0.1 s)
Loading cached output from `/root/.phdcache/runsim/2023-02-07__AdEx_Nto1__N=1600__T=600.jld2` … done (0.3 s)
Loading cached output from `/root/.phdcache/runsim/2023-02-07__AdEx_Nto1__N=6500__T=600.jld2` … done (0.8 s)
sims = first.(simruns)
inps = last.(simruns);
Base.summarysize(simruns[6]) / GB
0.523
Disentangle¶
spiketimes(input::Nto1Input) = input.train.spiketimes;
vrec(s::Simulation{<:Nto1System}) = s.rec.v;
/ end same
Plot¶
Let’s see what this AdEx guy looks like.
plot1(i) = begin
s = sims[i]
Nt = s.stepcounter.N
t = linspace(0, sim_duration, Nt)
plotsig(t, vrec(s) / mV; tlim=[0, 1seconds])
end
plot1(1)
plot1(3)
plot1(5)
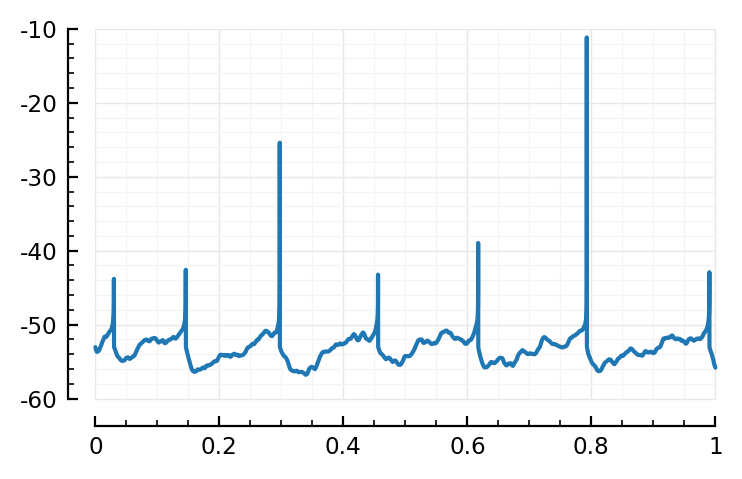
plot1(6);
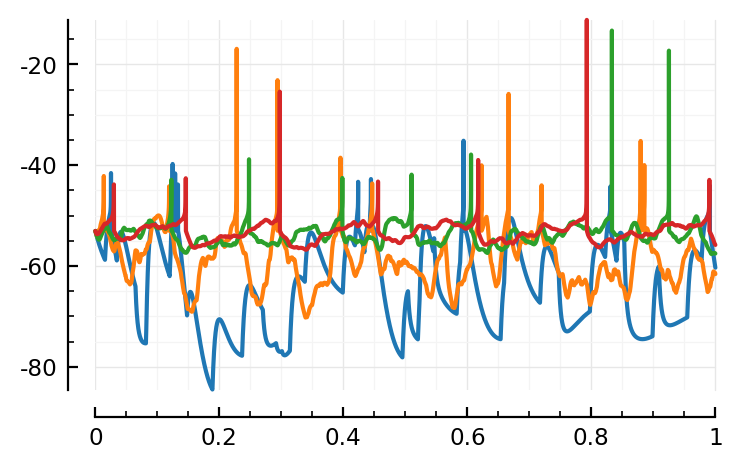
plot1(6);
Now to recreate the plot of https://tfiers.github.io/phd/nb/2022-10-24__Nto1_with_fixed_lognormal_inputs.html#plot
We’ll add fake spikes up to our previous’ spike cutoff.
i = 6
s = sims[i]
v = copy(s.rec.v);
v[to_timesteps.(s.rec.spiketimes, Δt)] .= 40mV
Nt = s.stepcounter.N
t = linspace(0, sim_duration, Nt)
plotsig(t, v / mV; tlim=[0, 10seconds]);
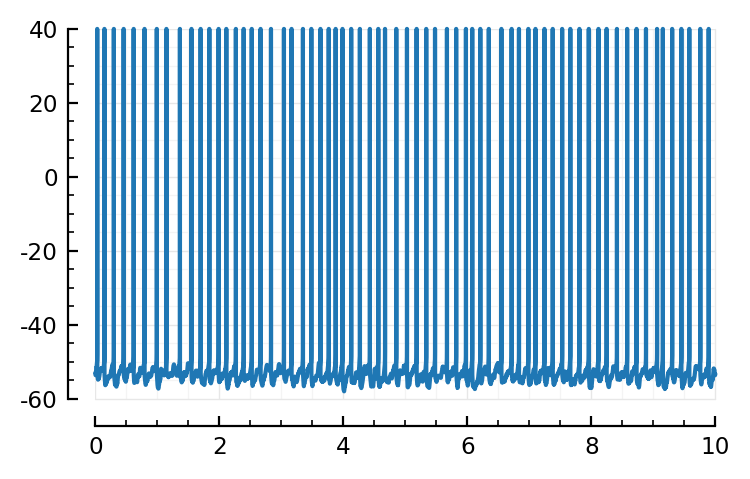
Conntest pooled windows - linear regress 10 ms¶
i = 6
N = Ns[i]
6500
inp = inps[i];
Nₑ = inp.Nₑ
5200
include("/root/phd/nb/2023-02-07__[input-linefit-wins].jl");
wins = windows(6, 1);
X, y = build_Xy(wins);
ts = @view X[:,2]
sel = 1:10000
Sciplotlib.plot(ts[sel]*Δt/ms, y[sel]/mV, ".", alpha=0.1);
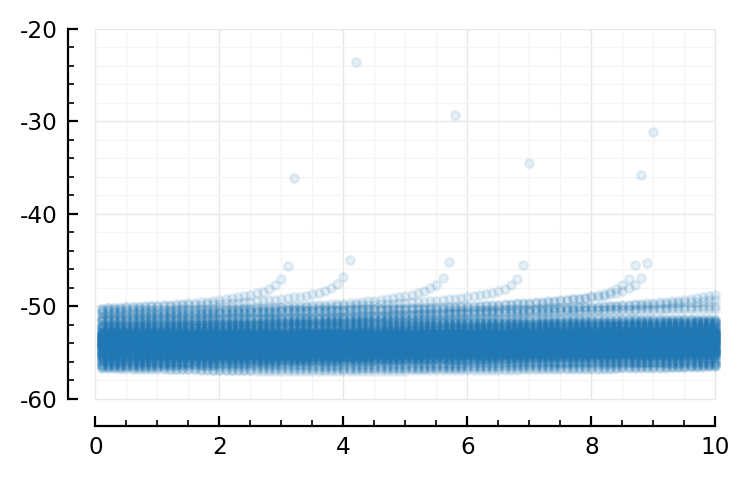
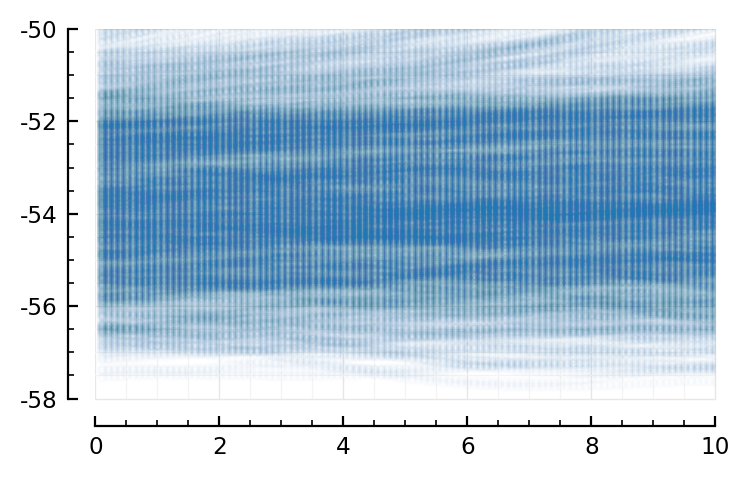
So we still see our ramp-ups; but much less so.
(compare, https://tfiers.github.io/phd/nb/2023-01-19__Fit-a-line.html#plot-some-windows)
Now the zoom-in
Ny = length(y)
280500
(only 0.3M datapoints here; but in the other nb we used our highest-firing).
sel = 1:100_000
Sciplotlib.plot(
ts[sel]*Δt/ms,
y[sel]/mV,
".";
alpha = 0.01,
ylim = [-58, -50], # mV
clip_on = true,
);
inh_neurons = Nₑ+1:N;
spiketimes(i::Int) = spiketimes(inp.inputs[i]);
shuffle_sources = sample(1:N, 100, replace=true)
real_spiketrains = spiketimes.(1:N);
unconnected_trains = shuffle_ISIs.(spiketimes.(shuffle_sources));
all_spiketrains = [real_spiketrains; unconnected_trains];
using Base.Threads: @threads
Nrows = length(all_spiketrains)
6600
(Below calc takes 3’25 on laptop, 7 threads)
makerows() = begin
rows = Vector(undef, Nrows)
p = Progress(Nrows)
@threads for r in 1:Nrows
rows[r] = makerow(r)
next!(p)
end
return rows
end;
rows = cached(makerows, (), key=cachekey(N));
Loading cached output from `/root/.phdcache/makerows/2023-02-07__AdEx_Nto1__N=6500__T=600.jld2` … done (0.1 s)
df = DataFrame(rows)
disp(df, 5) # (huh, disp no work no more here)
| Row | conntype | slope | pval | predtype |
|---|---|---|---|---|
| Symbol | Float64 | Float64 | Symbol | |
| 1 | exc | -0.28 | 0.813 | unconn |
| 2 | exc | 0.828 | 0.584 | unconn |
| 3 | exc | 3.78 | 1.44E-05 | exc |
| 4 | exc | 5.42 | 0.00335 | unconn |
| 5 | exc | 1.85 | 0.0458 | unconn |
| 6 | exc | 4.7 | 9.53E-05 | exc |
| 7 | exc | -1.3 | 0.26 | unconn |
| 8 | exc | 8.08 | 0.000785 | exc |
| 9 | exc | 1.65 | 0.00208 | unconn |
| 10 | exc | 8.21 | 6.55E-05 | exc |
| 11 | exc | 4.46 | 0.00175 | unconn |
| 12 | exc | 3.78 | 0.000389 | exc |
| 13 | exc | 2.73 | 0.00868 | unconn |
| ⋮ | ⋮ | ⋮ | ⋮ | ⋮ |
| 6589 | unconn | 3.52 | 0.0157 | unconn |
| 6590 | unconn | 0.62 | 0.428 | unconn |
| 6591 | unconn | 0.275 | 0.818 | unconn |
| 6592 | unconn | -0.95 | 0.391 | unconn |
| 6593 | unconn | -0.67 | 0.118 | unconn |
| 6594 | unconn | 4.07 | 0.002 | unconn |
| 6595 | unconn | 0.0647 | 0.961 | unconn |
| 6596 | unconn | 2.65 | 0.171 | unconn |
| 6597 | unconn | -4.21 | 9.3E-06 | inh |
| 6598 | unconn | -1.86 | 0.221 | unconn |
| 6599 | unconn | 0.427 | 0.612 | unconn |
| 6600 | unconn | -2.7 | 0.0612 | unconn |
perftable(df)
| Tested connections: 6600 | ||||||
|---|---|---|---|---|---|---|
| ┌─────── | Real type | ───────┐ | Precision | |||
unconn | exc | inh | ||||
| ┌ | unconn | 86 | 3712 | 730 | 2% | |
| Predicted type | exc | 11 | 1326 | 10 | 98% | |
| └ | inh | 3 | 161 | 561 | 77% | |
| Sensitivity | 86% | 26% | 43% |
At this arbitrary ‘α’ = 0.001:
FPR: 14%
TPRₑ: 26%
TPRᵢ: 43%
Comparing with the results with the Izh neuron:
(https://tfiers.github.io/phd/nb/2023-01-19__Fit-a-line.html#proper-eval)\
FPR: 34%
TPRₑ: 24%
TPRᵢ: 37%
So, that seems like a def increase :)
Now with lower FPR / lower α¶
Nrows = length(all_spiketrains)
# Nrows = 20
6600
α=0.0001;
rows2 = Vector(undef, Nrows)
p = Progress(Nrows)
@threads for r in 1:Nrows
rows2[r] = makerow(r; α)
next!(p)
end;
df2 = DataFrame(rows2)
perftable(df2)
Progress: 100%|█████████████████████████████████████████| Time: 0:04:54Progress: 73%|█████████████████████████████▊ | ETA: 0:01:21
| Tested connections: 6600 | ||||||
|---|---|---|---|---|---|---|
| ┌─────── | Real type | ───────┐ | Precision | |||
unconn | exc | inh | ||||
| ┌ | unconn | 84 | 4143 | 857 | 2% | |
| Predicted type | exc | 3 | 965 | 2 | 99% | |
| └ | inh | 13 | 91 | 442 | 81% | |
| Sensitivity | 84% | 19% | 34% |
(FPR 16%)
Even lowerr¶
It’s dumb to recalculate; we have the p-values.
(Plus, there’s some memory thing it seems: process dies halfway here).
update_predtype(row::DataFrameRow; α) = begin
if row.pval < α
predtype = (row.slope > 0 ? :exc : :inh)
else
predtype = :unconn
end
row.predtype = predtype
end;
df3 = deepcopy(df)
foreach(row -> update_predtype(row, α = 0.0000008), eachrow(df3))
perftable(df3)
| Tested connections: 6600 | ||||||
|---|---|---|---|---|---|---|
| ┌─────── | Real type | ───────┐ | Precision | |||
unconn | exc | inh | ||||
| ┌ | unconn | 95 | 4666 | 1019 | 2% | |
| Predicted type | exc | 5 | 501 | 1 | 99% | |
| └ | inh | 0 | 32 | 281 | 90% | |
| Sensitivity | 95% | 10% | 22% |
:D
Conntest STA¶
winsize = 1000
calcSTA(sim, spiketimes) =
calc_STA(vrec(sim), spiketimes, sim.Δt, winsize);
# @code_warntype calc_STA(vrec(s), st1, s.Δt, winsize)
# all good
Cache STA calc¶
using Base.Threads: @threads
function calc_STA_and_shufs(spiketimes, sim)
realSTA = calcSTA(sim, spiketimes)
shufs = [
calcSTA(sim, shuffle_ISIs(spiketimes))
for _ in 1:100
]
(; realSTA, shufs)
end
"calc_all_STAs_and_shufs"
function calc_all_STAz(inputs, sim)
f(input) = calc_STA_and_shufs(spiketimes(input), sim)
N = length(inputs)
res = Vector(undef, N)
p = Progress(N)
# @threads for i in 1:N
for i in 1:N
res[i] = f(inputs[i])
next!(p)
end
res
end
calc_all_STAz(simrun) = calc_all_STAz(unpakk(simrun)...);
unpakk(simrun) = (; simrun.input.inputs, simrun.sim);
# out = calc_all_STAz(simruns[1])
# print(Base.summary(out))
calc_all_cached(i) = cached(calc_all_STAz, [simruns[i]], key=cachekey(Ns[i]))
out = []
for i in eachindex(simruns)
push!(out, calc_all_cached(i))
end;
Loading cached output from `/root/.phdcache/calc_all_STAz/2023-02-07__AdEx_Nto1__N=5__T=600.jld2` … done (0.3 s)
Loading cached output from `/root/.phdcache/calc_all_STAz/2023-02-07__AdEx_Nto1__N=20__T=600.jld2` … done
Loading cached output from `/root/.phdcache/calc_all_STAz/2023-02-07__AdEx_Nto1__N=100__T=600.jld2` … done (0.2 s)
Loading cached output from `/root/.phdcache/calc_all_STAz/2023-02-07__AdEx_Nto1__N=400__T=600.jld2` … done (1.0 s)
Loading cached output from `/root/.phdcache/calc_all_STAz/2023-02-07__AdEx_Nto1__N=1600__T=600.jld2` … done (4.8 s)
Progress: 55%|██████████████████████▍ | ETA: 0:15:39
conntype_vec(i) = begin
sim, inp = simruns[i]
Nₑ = inp.Nₑ
N = Ns[i]
conntype = Vector{Symbol}(undef, N);
conntype[1:Nₑ] .= :exc
conntype[Nₑ+1:end] .= :inh
conntype
end;
conntestresults(i, teststat = ptp_test; α = 0.05) = begin
f((sta, shufs)) = test_conn(teststat, sta, shufs; α)
res = @showprogress map(f, out[i])
df = DataFrame(res)
df[!, :conntype] = conntype_vec(i)
df
end;
# conntestresults(1)
using Sciplotlib: plot
spikerate_(spiketimes) = length(spiketimes) / sim_duration;
spikerate_(inp::Nto1Input) = spikerate_(spiketimes(inp));
firing_rates(i) = spikerate_.(spiketimes.(inps[i].inputs));