2022-07-26 • Multiseed
Contents
2022-07-26 • Multiseed¶
Params¶
Based on Roxin; same as previous nb’s.
d = 6
ps = [
get_params(
duration = 10minutes,
p_conn = 0.04,
g_EE = 1 / d,
g_EI = 18 / d,
g_IE = 36 / d,
g_II = 31 / d,
ext_current = Normal(-0.5 * pA/√seconds, 5 * pA/√seconds),
E_inh = -80 * mV,
record_v = [1:40; 801:810;];
rngseed,
)
for rngseed in [VoltoMapSim.default_rngseed, 0]
];
Run sim¶
ss = [cached(sim, [p.sim]) for p in ps];
ss = [augment_simdata(s, p) for (s,p) in zip(ss,ps)];
Eval conntest perf for all v recorded¶
recorded = p[1].sim.network.record_v;
# using Base.Threads
detrates = [Dict(), Dict()]
for i in [1,2]
# @threads for m in recorded
for m in recorded
ii = get_input_info(m, ss[i], ps[i]);
perf = cached(evaluate_conntest_perf, [ii.v, ii.spiketrains, ps[i]], key=[ps[i], m])
detrates[i][m] = perf.detection_rates
end
end;
Plot distributions¶
using PyPlot
using VoltoMapSim.Plot
exc_rec = [1:40;]
inh_rec = [801:810;];
ydistplot = VoltoMapSim.Plot.ydistplot;
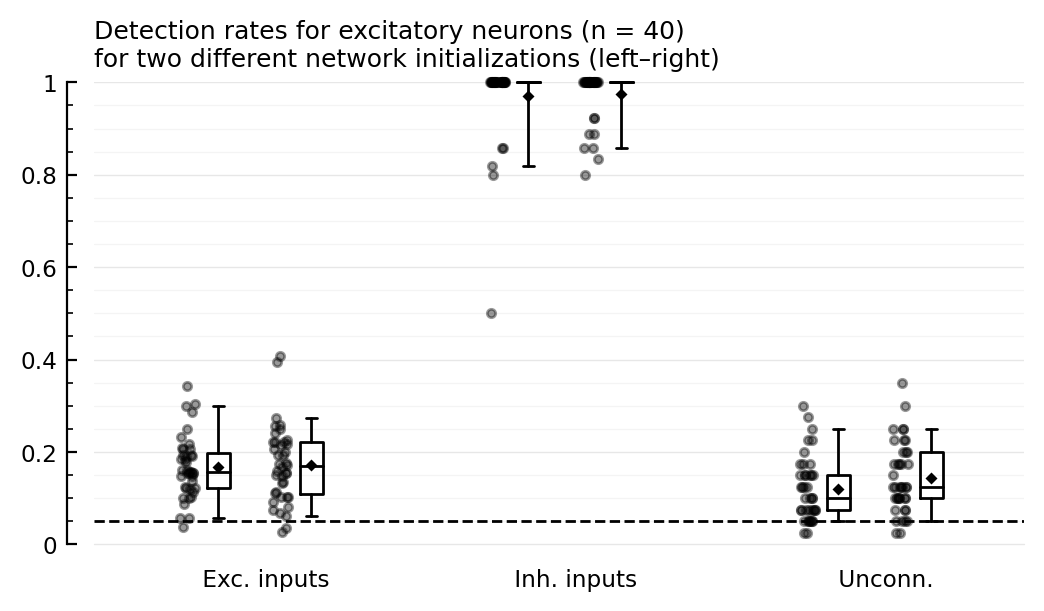
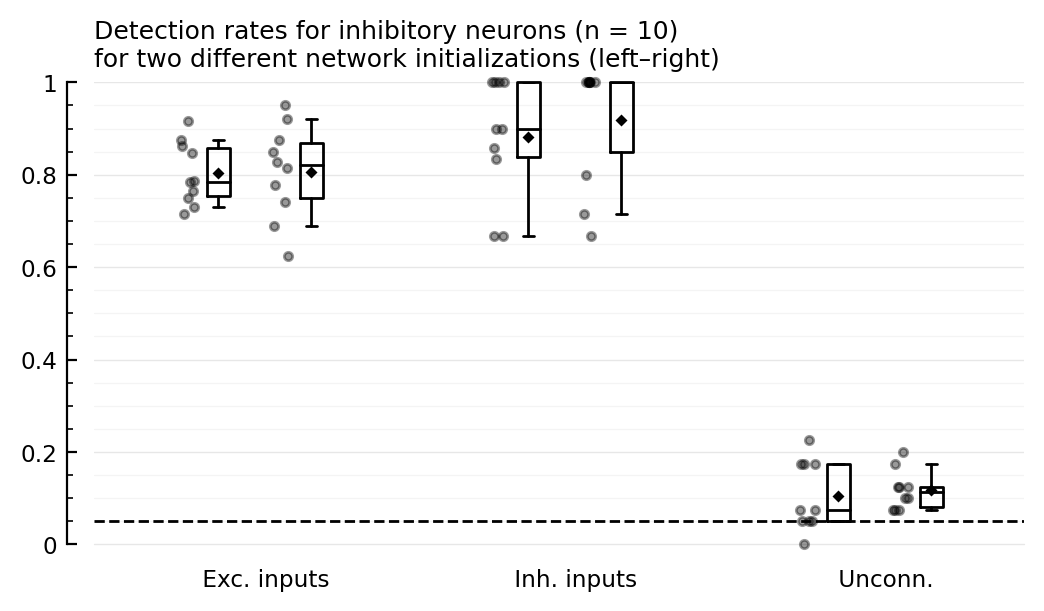
function detplot(coll, name)
N = length(coll)
fill = " "^17
ax = ydistplot(
fill*"Exc. inputs" => [detrates[1][n].TPR_exc for n in coll],
"" => [detrates[2][n].TPR_exc for n in coll],
fill*"Inh. inputs" => [detrates[1][n].TPR_inh for n in coll],
"" => [detrates[2][n].TPR_inh for n in coll],
fill*"Unconn." => [detrates[1][n].FPR for n in coll],
"" => [detrates[2][n].FPR for n in coll],
xpos = [1.2, 1.8, 3.2, 3.8, 5.2, 5.8],
figsize = (6,3),
ylim = [0,1],
hylabel = ("Detection rates for $(name) neurons (n = $N)\n"
* "for two different network initializations (left–right)"
),
)
add_α_line(ax, p[1].evaluation.α)
return nothing
end;
detplot(exc_rec, "excitatory")
detplot(inh_rec, "inhibitory")
Is there relation between exc and inh det performance?¶
exc_in = [detrates[1][n].TPR_exc for n in inh_rec]
inh_in = [detrates[1][n].TPR_inh for n in inh_rec]
fig,ax=plt.subplots(figsize=(2.6,2.6))
ax.plot(exc_in, inh_in, "k.", clip_on=false)
ax.set(xlim=[0,1], ylim=[0,1], aspect="equal")
set(ax, xlabel="E→I det rate", ylabel=("I→I det rate", :loc=>"top"));
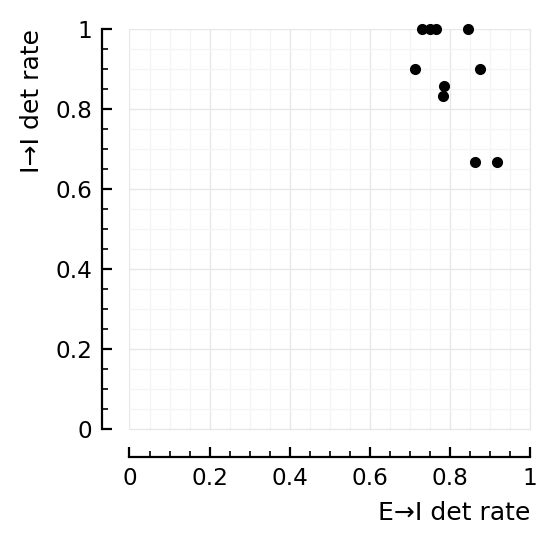
Seems not. Maybe anticorrelated but eh.
What about for exc postsyn¶
exc_in = [detrates[1][n].TPR_exc for n in exc_rec]
inh_in = [detrates[1][n].FPR for n in exc_rec]
fig,ax=plt.subplots(figsize=(2.6,2.6))
ax.plot(exc_in, inh_in, "k.", clip_on=false)
ax.set(xlim=[0,1], ylim=[0,1], aspect="equal")
set(ax, xlabel="E→E det rate", ylabel=("unconn→E det rate", :loc=>"top"));
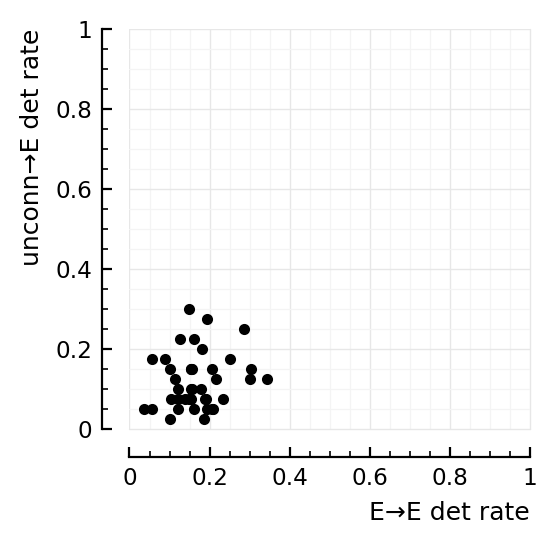
Eh