2022-07-23 • Record many neurons
Contents
2022-07-23 • Record many neurons¶
Params¶
Based on Roxin; same as previous nb’s.
d = 6
p = get_params(
duration = 10minutes,
#duration = 10seconds,
p_conn = 0.04,
g_EE = 1 / d,
g_EI = 18 / d,
g_IE = 36 / d,
g_II = 31 / d,
ext_current = Normal(-0.5 * pA/√seconds, 5 * pA/√seconds),
E_inh = -80 * mV,
record_v = [1:40; 801:810;],
);
Eval conntest perf for all v recorded¶
recorded = p.sim.network.record_v;
# using Base.Threads
detrates = Dict()
# @threads for m in recorded
for m in recorded
ii = get_input_info(m, s, p);
perf = cached(evaluate_conntest_perf, [ii.v, ii.spiketrains, p], key=[p, m])
detrates[m] = perf.detection_rates
end;
Connection tests take ~1:20 per neuron. So for 50 neurons → 66’.
Testing can happen multithreaded. So we’ll set the JULIA_NUM_THREADS env var (to one less than the num logical cores).
~~Ah seems you can use all cores, as it doesn’t utilize fully, so there’s left for you.~~ Nope, using all cores clogs pc.
With the threading it was sometimes up to 7:30 per neuron..
Plot distributions¶
using PyPlot
using VoltoMapSim.Plot
exc_rec = [1:40;]
inh_rec = [801:810;];
function ydistplot(pairs...; setkw...)
labels = [p[1] for p in pairs]
datas = [p[2] for p in pairs]
fig, ax = plt.subplots()
for (x, ys) in enumerate(datas)
xs = x .- 0.15 .+ 0.1*rand(length(ys))
ax.plot(xs, ys, "k.", clip_on=false, alpha=0.4)
ax.boxplot(
ys, whis=(5,95), positions=[x+0.1], showfliers=false, showmeans=true,
medianprops=Dict(:color=>"black"),
meanprops=Dict(:marker=>"D", :ms=>3, :mfc=>"black", :mec=>"none")
)
end
N = length(labels)
set(ax; xtype=:categorical, xlim=[0.5, N+0.5], setkw...)
ax.set_xticks(1:N) # can't use `set` as that sets xticks
ax.set_xticklabels(labels)
return ax
end;
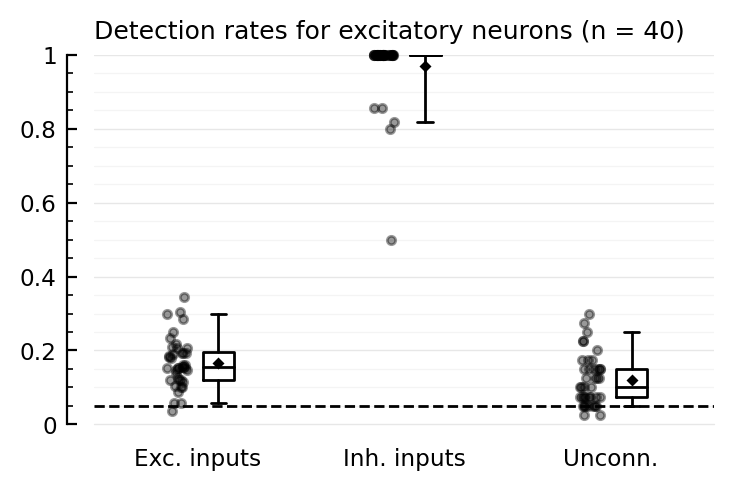
function detplot(coll, name)
N = length(coll)
ax = ydistplot(
"Exc. inputs" => [detrates[n].TPR_exc for n in coll],
"Inh. inputs" => [detrates[n].TPR_inh for n in coll],
"Unconn." => [detrates[n].FPR for n in coll],
ylim = [0,1],
hylabel = "Detection rates for $(name) neurons (n = $N)",
)
add_α_line(ax, p.evaluation.α)
return nothing
end;
detplot(exc_rec, "excitatory")
detplot(inh_rec, "inhibitory")
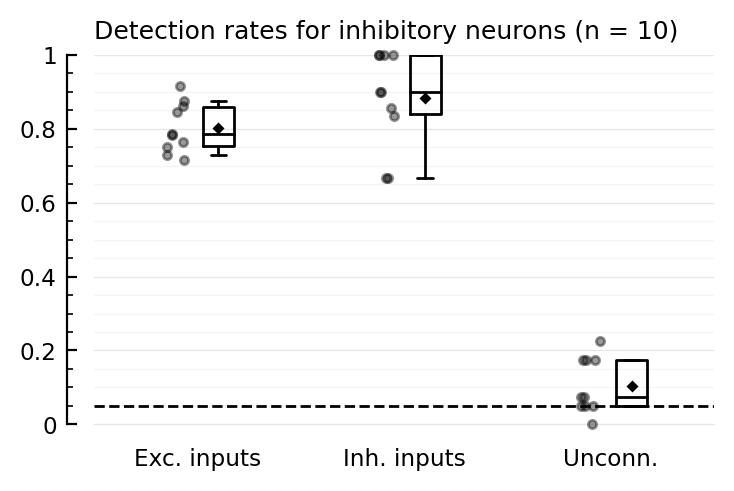
Whiskers are the 5 and 95 percentiles.