2020-11-30 • Speedup spikes & STA calc
Contents
2020-11-30 • Speedup spikes & STA calc¶
A notebook based on /notebooks/2020-11-27__permutation_test, in which we try to speed-up spike train generation & shufflin, and STA calculation.
Imports & time grid¶
from voltage_to_wiring_sim.support.notebook_init import *
Preloading:
- numpy … (0.11 s)
- matplotlib.pyplot … (0.23 s)
- numba … (0.30 s)
Importing from submodules (compiling numba functions) … ✔
Imported `np`, `mpl`, `plt`
Imported codebase (`voltage_to_wiring_sim`) as `v`
Imported `*` from `v.support.units`
Setup autoreload
v.print_reproducibility_info()
This cell was last run by tfiers on yoga
on Mon 30 Nov 2020, at 13:58 (UTC+0100).
Last git commit (Mon 30 Nov 2020, 13:44).
Uncommited changes to:
M codebase/voltage_to_wiring_sim/STA.py
M codebase/voltage_to_wiring_sim/__init__.py
M codebase/voltage_to_wiring_sim/spike_train.py
A notebooks/2020-11-11__unitlib.ipynb
?? notebooks/2020-11-30__speedup.ipynb
tg = v.TimeGrid(T=10*minute, dt=0.1*ms);
Generate VI signal¶
Spike trains¶
N_in = 30
p_connected = 0.5
N_connected = round(N_in * p_connected)
N_unconnected = N_in - N_connected
15
f_spike = 20 * Hz;
gen_st = v.generate_Poisson_spike_train
v.fix_rng_seed()
%%time
spike_trains_connected = [gen_st(tg, f_spike) for _ in range(N_connected)]
spike_trains_unconnected = [gen_st(tg, f_spike) for _ in range(N_unconnected)];
Wall time: 1.77 s
all_spike_trains = spike_trains_connected + spike_trains_unconnected;
time_slice = 1 * minute + np.array([0, 1]) * second
slice_indices = np.round(time_slice / tg.dt).astype(int)
i_slice = slice(*slice_indices)
t_slice = tg.t[i_slice]
array([60, 60, 60, ..., 61, 61, 61])
def plot_spike_train_excerpt(spike_train):
return v.spike_train.plot(t_slice, spike_train[i_slice])
plot_spike_train_excerpt(all_spike_trains[0]);
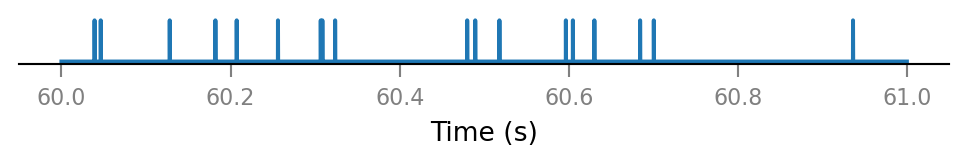
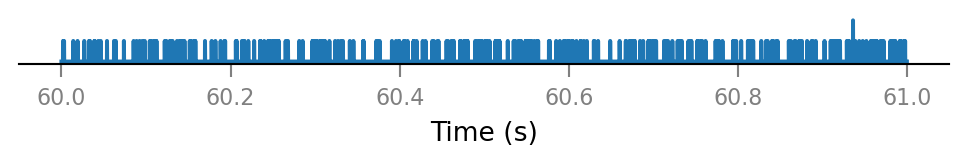
all_incoming_spikes = sum(spike_trains_connected)
v.spike_train.plot(t_slice, all_incoming_spikes[i_slice]);
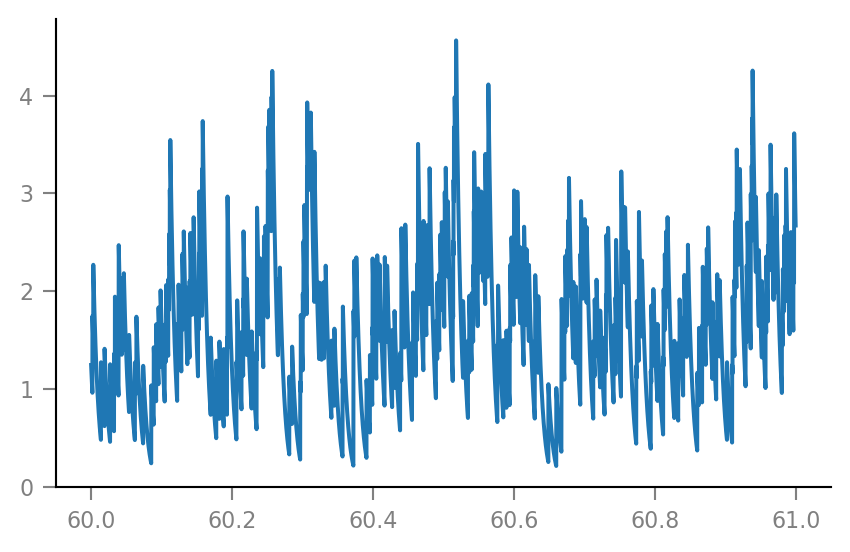
Synaptic conductance¶
Δg_syn = 0.8 * nS
τ_syn = 7 * ms;
g_syn = v.calc_synaptic_conductance(
tg, all_incoming_spikes, Δg_syn, τ_syn)
plt.plot(t_slice, g_syn[i_slice] / nS);
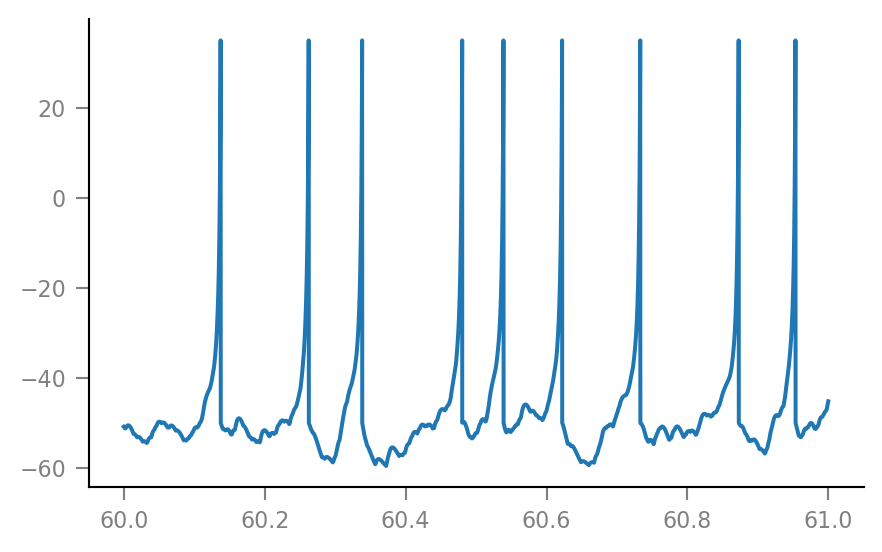
Membrane voltage¶
params = v.params.cortical_RS
v.pprint(params)
IzhikevichParams
----------------
C = 1e-10
k = 7e-07
v_r = -0.06
v_t = -0.04
v_peak = 0.035
a = 30.0
b = -2e-09
c = -0.05
d = 1e-10
v_syn = 0.0
%%time
sim = v.simulate_izh_neuron(tg, params, g_syn)
Wall time: 240 ms
plt.plot(t_slice, sim.V_m[i_slice] / mV);
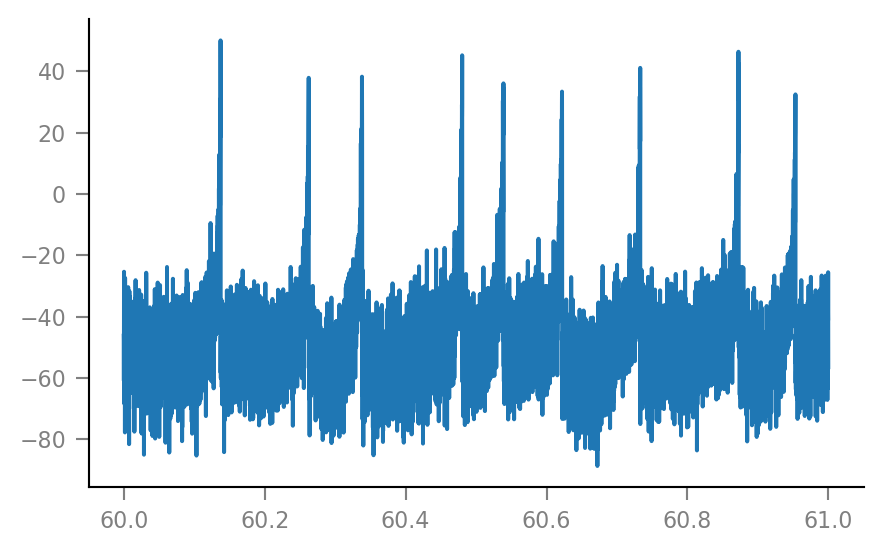
Noise¶
Vm_noisy = v.add_VI_noise(sim.V_m, params)
plt.plot(t_slice, Vm_noisy[i_slice] / mV);
Shuffle one spike train¶
We choose one spike train / incoming neuron, and will test the hypothesis that this neuron is connected to the simulated neuron.
H0 = neuron of incoming spike train is not connected to simulated neuron
H1 = they are connected
spike_train = all_spike_trains[0];
We will shuffle the real spike train \(S\) times to generate \(S\) ‘fake’ spike trains, which have the same number of spikes but at random times.
S = 100;
%%time
shuffled_spike_trains = []
for i in range(S):
x = spike_train.copy()
np.random.shuffle(x) # in-place
shuffled_spike_trains.append(x)
Wall time: 14 s
So this is slow. Speedup by shuffling not the entire time-series, but only the inter-spike intervals (as suggested by Mark).
def ISI(spike_train):
'''
Inter-spike intervals, in number of samples.
First element is not strictly an ISI, but rather the time of
the first spike after beginning of the time-series.
'''
spike_indices = v.spike_train_to_indices(spike_train)
ISI = np.diff(spike_indices, prepend=[0])
return ISI
ISI_original = ISI(spike_train)
array([757, 64, 249, ..., 221, 53, 881], dtype=int64)
def ISI_to_spike_indices(ISI):
return np.cumsum(ISI)
%%time
shuffled_spike_trains_ = []
for i in range(S):
ISI_shuffled = ISI_original.copy()
np.random.shuffle(ISI_shuffled) # in-place
spike_indices_shuffled = ISI_to_spike_indices(ISI_shuffled)
spike_train_shuffled = v.spike_train_from_indices(spike_indices_shuffled, tg)
shuffled_spike_trains_.append(spike_train_shuffled)
Wall time: 708 ms
that’s 20x speedup :)
%%time
shuffled_spike_ix_ = []
for i in range(S):
ISI_shuffled = ISI_original.copy()
np.random.shuffle(ISI_shuffled) # in-place
spike_indices_shuffled = ISI_to_spike_indices(ISI_shuffled)
shuffled_spike_ix_.append(spike_indices_shuffled)
Wall time: 34 ms
Not using full spike trains (but only indices) yields another 20x speedup.
Inspect a few shuffled spike trains.
for i in range(3):
plot_spike_train_excerpt(shuffled_spike_trains[i]);
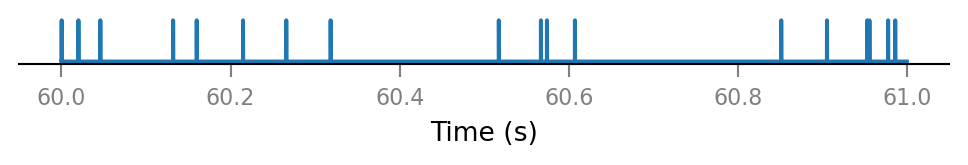
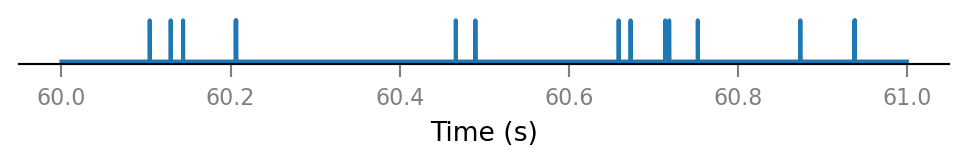
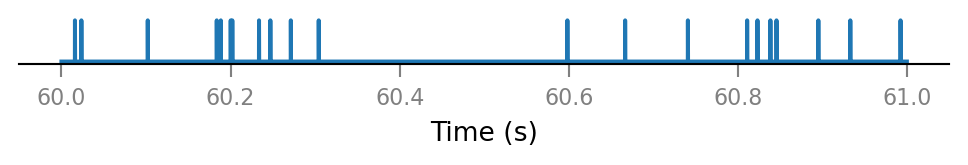
for i in range(3):
plot_spike_train_excerpt(shuffled_spike_trains_[i]);
Real spike train again:
plot_spike_train_excerpt(spike_train);
Same number of spikes in all:
def num_spikes(spike_train):
spike_indices = v.spike_train_to_indices(spike_train)
return len(spike_indices)
for i in range(3):
print(num_spikes(shuffled_spike_trains[i]))
12020
12020
12020
for i in range(3):
print(num_spikes(shuffled_spike_trains_[i]))
12020
12020
12020
num_spikes(spike_train)
12020
len(shuffled_spike_ix_[0])
12020
Spiking frequency of the chosen presynaptic neuron:
num_spikes(spike_train) / tg.T / Hz
20.03
That’s to spec.
Spike-triggered windowing & averaging¶
For each spike train (original and shuffleds), we extract spike-triggered windows from the simulated voltage imaging signal, and average those windows.
window_length = 100 * ms;
window_tg = v.TimeGrid(window_length, tg.dt);
def calc_STA(spike_train):
return v.calculate_STA(Vm_noisy, spike_train, tg, window_tg)
original_STA = calc_STA(spike_train);
%%time
shuffled_STAs = []
for train in shuffled_spike_trains:
shuffled_STAs.append(calc_STA(train))
Wall time: 8.36 s
Now with numba JIT’ted make_windows:
%%prun -D temp.profile
shuffled_STAs = []
for train in shuffled_spike_trains:
shuffled_STAs.append(calc_STA(train))
*** Profile stats marshalled to file 'temp.profile'.
(Time was like 6.4 seconds).
Now with creating 2D ndarray directly, instead of Python list of arrays:
(pure python:)
%%time
shuffled_STAs = []
for train in shuffled_spike_trains:
shuffled_STAs.append(calc_STA(train))
Wall time: 7.75 s
(njit):
%%time
shuffled_STAs = []
for train in shuffled_spike_trains:
shuffled_STAs.append(calc_STA(train))
Wall time: 5.11 s
%%prun -D temp.profile2
shuffled_STAs = []
for train in shuffled_spike_trains:
shuffled_STAs.append(calc_STA(train))
*** Profile stats marshalled to file 'temp.profile2'.
Don’t create windows list; calc mean (sum) immediately:
%%time
shuffled_STAs = []
for train in shuffled_spike_trains:
shuffled_STAs.append(calc_STA(train))
Wall time: 725 ms
%%prun -D temp.profile3
shuffled_STAs = []
for train in shuffled_spike_trains:
shuffled_STAs.append(calc_STA(train))
*** Profile stats marshalled to file 'temp.profile3'.
Ok, this is good improvement (from 8 to 0.8 seconds, i.e. 10x).
If we now also get rid of full spike trains and replace them with spike indices, we’re good.
(0.4 s in njitted direct _calc_STA, 0.3 s in np.nonzero (i.e. v.get_spike_indices).
Repro info¶
v.print_reproducibility_info(verbose=True)
This cell was last run by tfiers on yoga
on Mon 30 Nov 2020, at 15:13 (UTC+0100).
Last git commit (Mon 30 Nov 2020, 14:39).
Uncommited changes to:
M codebase/voltage_to_wiring_sim/STA.py
A notebooks/2020-11-11__unitlib.ipynb
?? notebooks/2020-11-30__speedup.ipynb
?? notebooks/temp.profile
?? notebooks/temp.profile2
?? notebooks/temp.profile3
Platform:
Windows-10
CPython 3.8.3 (C:\conda\python.exe)
Intel(R) Core(TM) i7-10510U CPU @ 1.80GHz
Dependencies of voltage_to_wiring_sim and their installed versions:
numpy 1.19.2
matplotlib 3.3.2
numba 0.51.2
seaborn 0.10.1
scipy 1.5.2
scikit-learn 0.23.2
preload 2.1
py-cpuinfo 7.0.0