2023-08-30__STA_examples
Contents
2023-08-30__STA_examples¶
(Result of workflow speed (& ergonomy) tests: full Julia (no Python hybrid))
So, for all 10 Ns;
For 10 diff seeds;
for both exc, inh, and unconn;
we conntest (maximum) 100 input spike trains.
(Each test is comprised of calculating 101 STAs: one real and the rest with shuffled ISIs).
From the prev nb (https://tfiers.github.io/phd/nb/2023-08-16__STA_conntest_pyjulia.html), we found we’d take a shorter window, so that ‘area over start’ measure (to determine if exc or inh) is correct.
But ok, it’s good to show that in thesis.
So, we repeat an example STA plot here.
for full N ofc.
include("lib/Nto1.jl")
using Revise … ✔ (0.3 s)
using Units, Nto1AdEx, ConnectionTests, ConnTestEval …
[ Info: Precompiling Nto1AdEx [368485ca-63bc-4029-854f-349d2205662c]
✔ (1.5 s)
using StatsBase … ✔ (0.2 s)
N = 6500;
duration = 10minutes
600
@time sim = Nto1AdEx.sim(N, duration);
2.994944 seconds (2.14 M allocations: 1.027 GiB, 2.34% gc time, 42.14% compilation time)
(1st run: 2.5 secs, 27% compilation time).
We want our input spiketrains sorted: the highest spikers first.
And split exc/inh, too.
include("lib/df.jl")
using DataFrames … ✔ (0.7 s)
exc_inputs = highest_firing(excitatory_inputs(sim))
tabulate(trains) = DataFrame(
"# input spikes" => num_spikes.(trains),
"spike rate (Hz)" => spikerate.(trains)
)
tabulate(exc_inputs)
| Row | # input spikes | spike rate (Hz) |
|---|---|---|
| Int64 | Float64 | |
| 1 | 58522 | 97.5 |
| 2 | 35312 | 58.9 |
| 3 | 24428 | 40.7 |
| 4 | 20653 | 34.4 |
| 5 | 18898 | 31.5 |
| ⋮ | ⋮ | ⋮ |
| 5196 | 194 | 0.323 |
| 5197 | 185 | 0.308 |
| 5198 | 175 | 0.292 |
| 5199 | 168 | 0.28 |
| 5200 | 123 | 0.205 |
inh_inputs = highest_firing(inhibitory_inputs(sim))
tabulate(inh_inputs)
| Row | # input spikes | spike rate (Hz) |
|---|---|---|
| Int64 | Float64 | |
| 1 | 21808 | 36.3 |
| 2 | 21054 | 35.1 |
| 3 | 16402 | 27.3 |
| 4 | 14905 | 24.8 |
| 5 | 14429 | 24 |
| ⋮ | ⋮ | ⋮ |
| 1296 | 234 | 0.39 |
| 1297 | 220 | 0.367 |
| 1298 | 218 | 0.363 |
| 1299 | 197 | 0.328 |
| 1300 | 140 | 0.233 |
( :) )
STA = calc_STA(sim.V, exc_inputs[1].times);
include("lib/plot.jl")
import PythonCall … ✔ (2 s)
import PythonPlot … ✔ (3.7 s)
using Sciplotlib … ✔ (0.5 s)
using PhDPlots … ✔
plotSTA(STA);
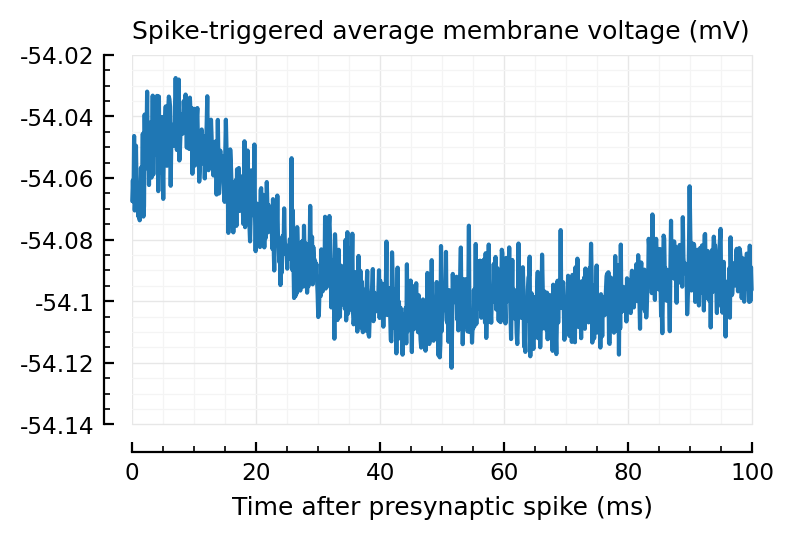
To compare with predicted PSP height (0.04 mV):
(maximum(STA) - first(STA)) / mV
0.0399
(Woah, that’s close)
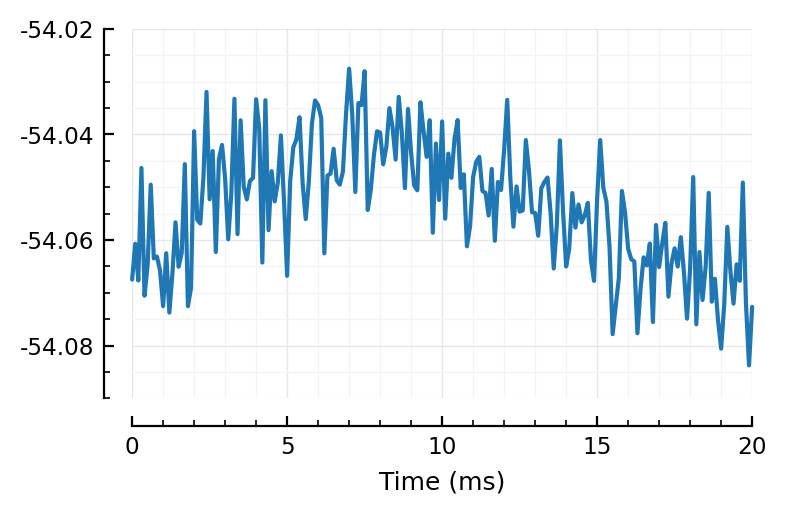
plotsig(STA/mV, [0,20], ms);
plotSTA_(train, sim=sim; kw...) = begin
nspikes = num_spikes(train)
EI = train ∈ exc_inputs ? "exc" : "inh"
label = "$nspikes spikes, $EI"
plotSTA(calc_STA(sim.V, train.times); label, kw...)
end
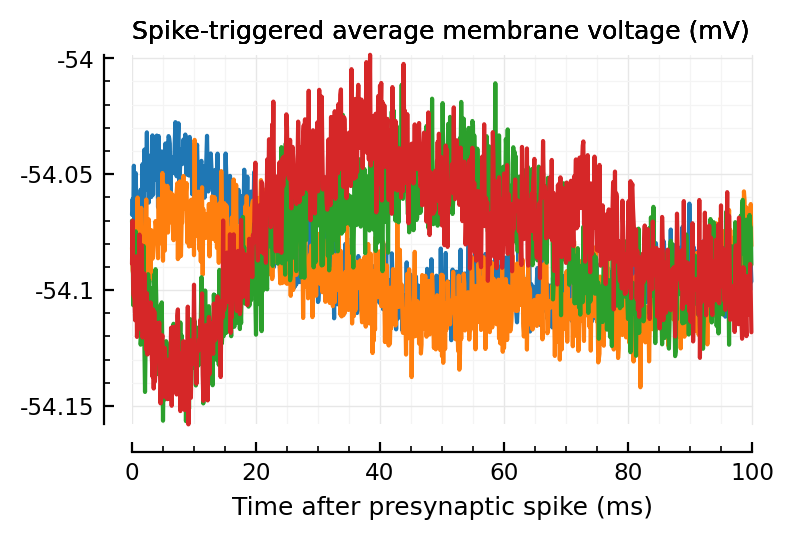
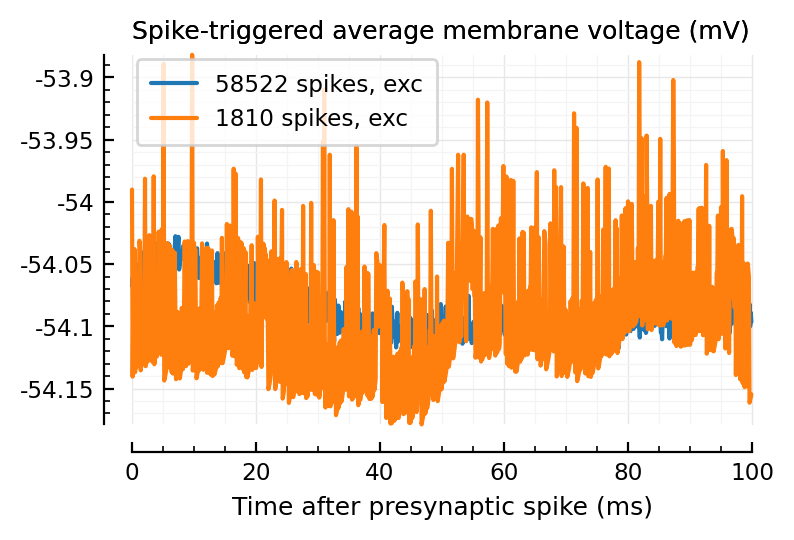
plotSTA_(exc_inputs[1]);
plotSTA_(exc_inputs[2]);
plotSTA_(inh_inputs[1]);
plotSTA_(inh_inputs[2]);
plotSTA_(exc_inputs[1]);
plotSTA_(exc_inputs[end]);
plt.legend();
mid = length(exc_inputs) ÷ 2
plotSTA_(exc_inputs[1]);
plotSTA_(exc_inputs[mid]);
plt.legend();
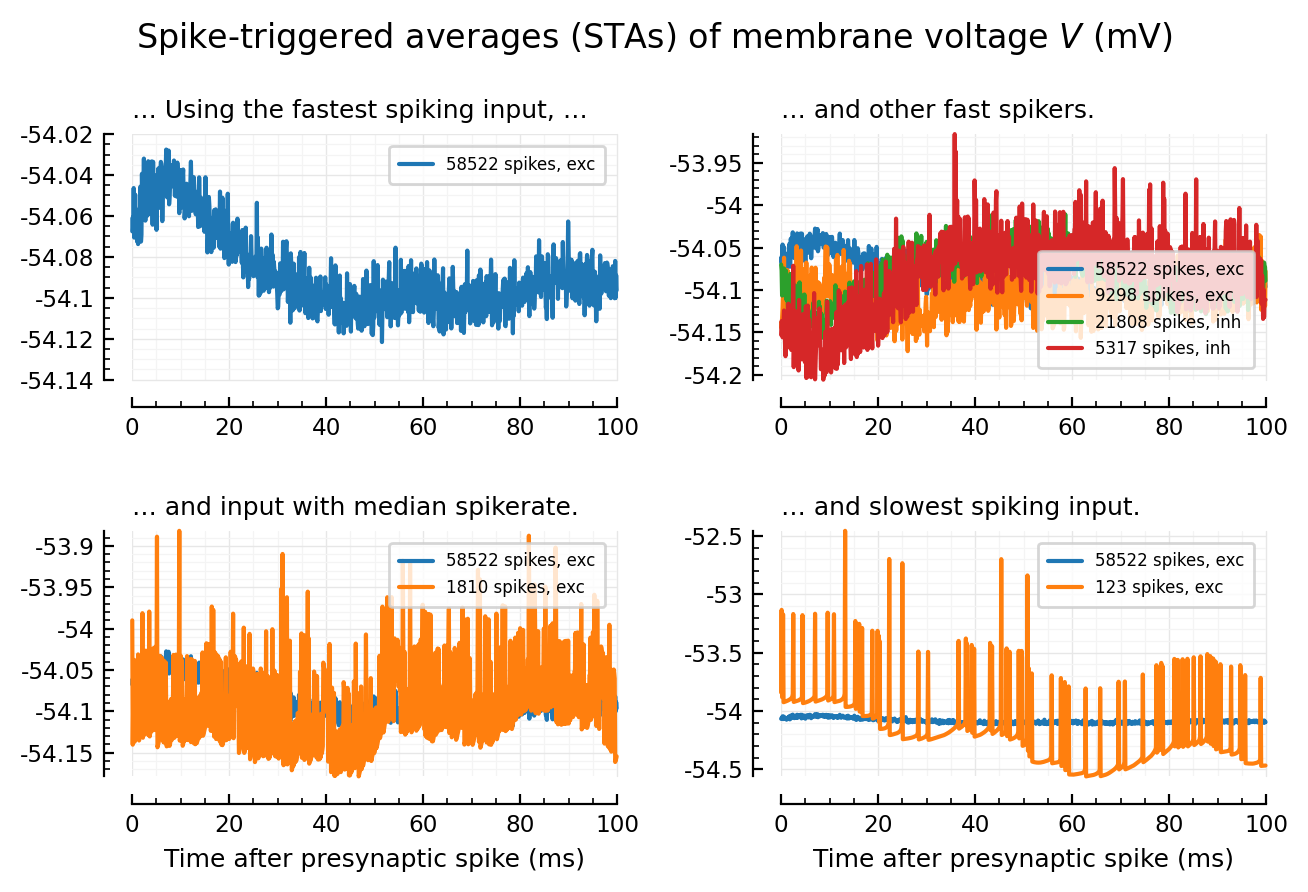
Four-panel plot¶
fig, axs = plt.subplots(nrows=2, ncols=2, figsize=(pw*0.8, mtw))
plotSTA_2(args...; hylabel=nothing, kw...) = plotSTA_(args...; hylabel, kw...)
addlegend(ax; kw...) = legend(ax, fontsize=6, borderaxespad=0.7; kw...)
plotSTA_2(exc_inputs[1], ax=axs[0,0], hylabel="… Using the fastest spiking input, …", xlabel=nothing);
addlegend(axs[0,0])
plotSTA_2(exc_inputs[1], ax=axs[0,1], hylabel="… and other fast spikers.", xlabel=nothing);
plotSTA_2(exc_inputs[100], ax=axs[0,1], xlabel=nothing)
plotSTA_2(inh_inputs[1], ax=axs[0,1], xlabel=nothing)
plotSTA_2(inh_inputs[100], ax=axs[0,1], xlabel=nothing)
addlegend(axs[0,1], loc="lower right")
plotSTA_2(exc_inputs[1], ax=axs[1,1], hylabel="… and slowest spiking input.");
plotSTA_2(exc_inputs[end], ax=axs[1,1]);
addlegend(axs[1,1])
plotSTA_2(exc_inputs[1], ax=axs[1,0], hylabel="… and input with median spikerate.");
plotSTA_2(exc_inputs[mid], ax=axs[1,0]);
addlegend(axs[1,0], loc="upper right")
plt.suptitle(L"Spike-triggered averages (STAs) of membrane voltage $V$ (mV)")
plt.tight_layout(h_pad=2);
# savefig_phd("example_STAs")
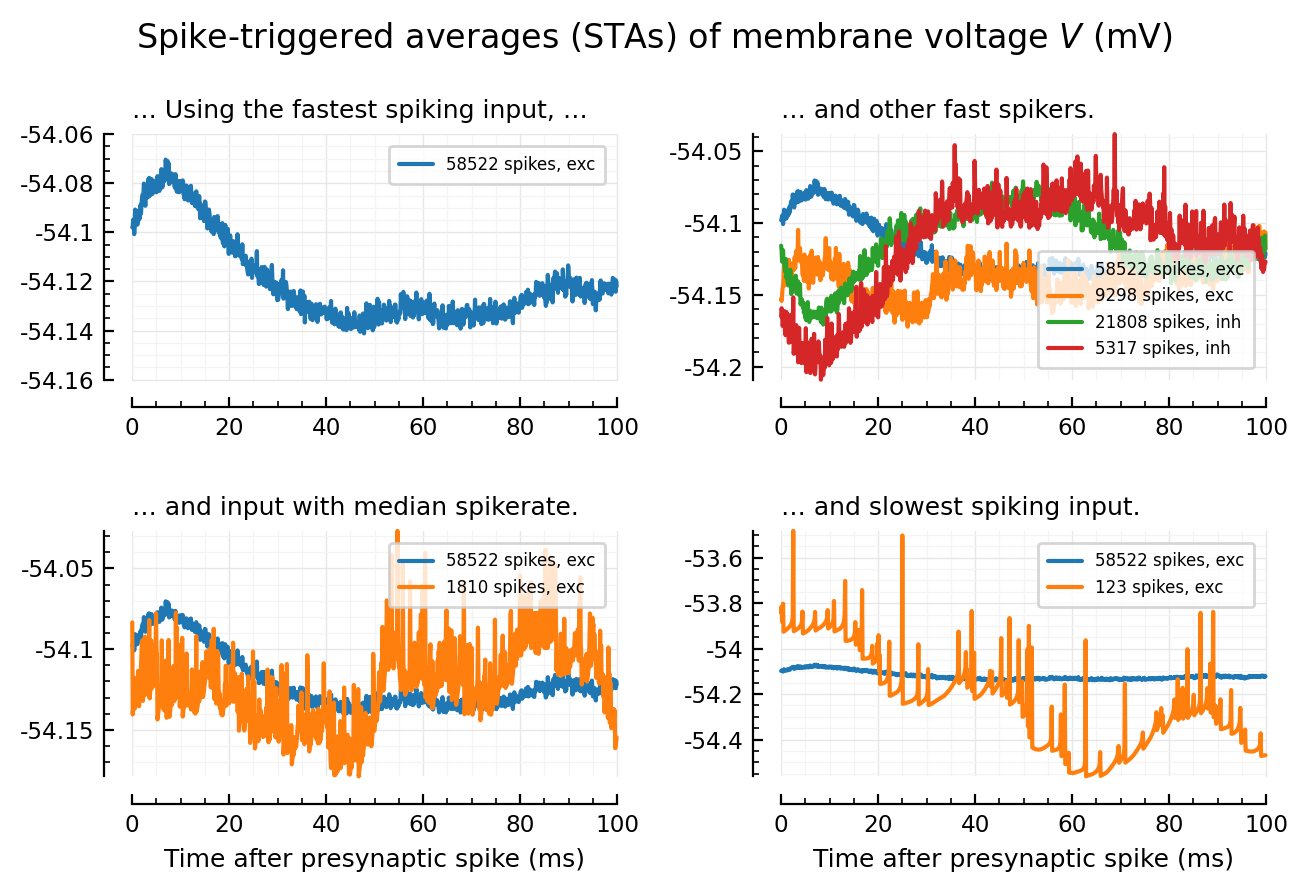
fig, axs = plt.subplots(nrows=2, ncols=2, figsize=(pw*0.8, mtw))
plotSTA_2(args...; hylabel=nothing, kw...) = plotSTA_(args...; hylabel, kw...)
addlegend(ax; kw...) = legend(ax, fontsize=6, borderaxespad=0.7; kw...)
plotSTA_2(exc_inputs[1], ax=axs[0,0], hylabel="… Using the fastest spiking input, …", xlabel=nothing);
addlegend(axs[0,0])
plotSTA_2(exc_inputs[1], ax=axs[0,1], hylabel="… and other fast spikers.", xlabel=nothing);
plotSTA_2(exc_inputs[100], ax=axs[0,1], xlabel=nothing)
plotSTA_2(inh_inputs[1], ax=axs[0,1], xlabel=nothing)
plotSTA_2(inh_inputs[100], ax=axs[0,1], xlabel=nothing)
addlegend(axs[0,1], loc="lower right")
plotSTA_2(exc_inputs[1], ax=axs[1,1], hylabel="… and slowest spiking input.");
plotSTA_2(exc_inputs[end], ax=axs[1,1]);
addlegend(axs[1,1])
plotSTA_2(exc_inputs[1], ax=axs[1,0], hylabel="… and input with median spikerate.");
plotSTA_2(exc_inputs[mid], ax=axs[1,0]);
addlegend(axs[1,0], loc="upper right")
plt.suptitle(L"Spike-triggered averages (STAs) of membrane voltage $V$ (mV)")
plt.tight_layout(h_pad=2);
# savefig_phd("example_STAs")
Saved at `../thesis/figs/example_STAs.pdf`
(For colour in figure caption text):
cs = darken.(Sciplotlib.mplcolors, 0.87)
toRGBAtuple.(cs)[1:6]
6-element Vector{NTuple{4, FixedPointNumbers.N0f8}}:
(0.106N0f8, 0.408N0f8, 0.616N0f8, 1.0N0f8)
(0.871N0f8, 0.431N0f8, 0.047N0f8, 1.0N0f8)
(0.149N0f8, 0.545N0f8, 0.149N0f8, 1.0N0f8)
(0.729N0f8, 0.133N0f8, 0.137N0f8, 1.0N0f8)
(0.506N0f8, 0.353N0f8, 0.643N0f8, 1.0N0f8)
(0.478N0f8, 0.294N0f8, 0.255N0f8, 1.0N0f8)
Ceil spikes¶
sim_ceil = deepcopy(sim)
ceil_spikes!(sim_ceil.V, sim_ceil.spiketimes);
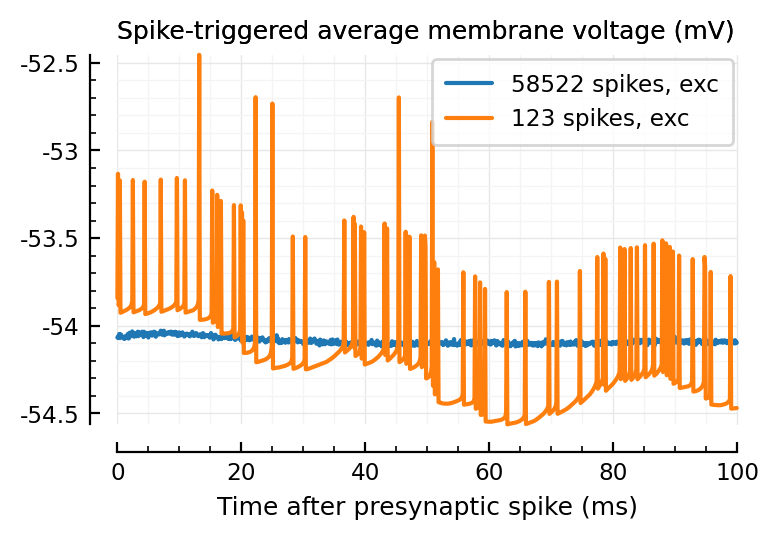
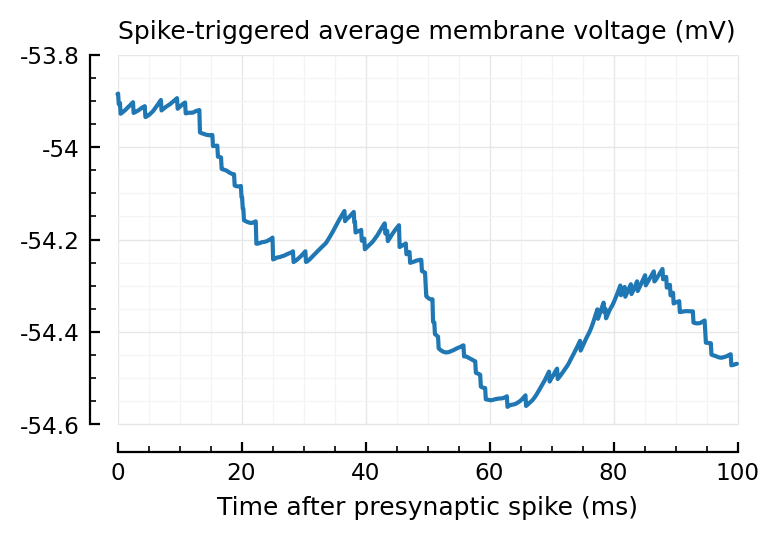
plotSTA_2(exc_inputs[1]);
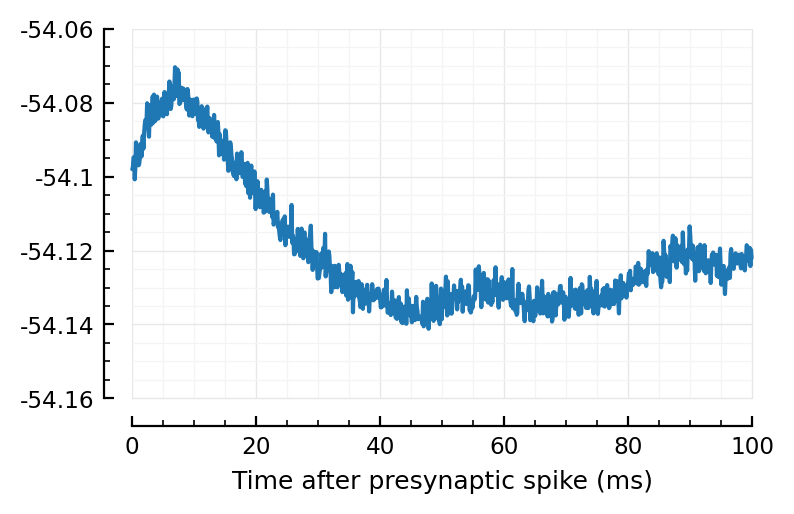
plotSTA_2(exc_inputs[1], sim_ceil);
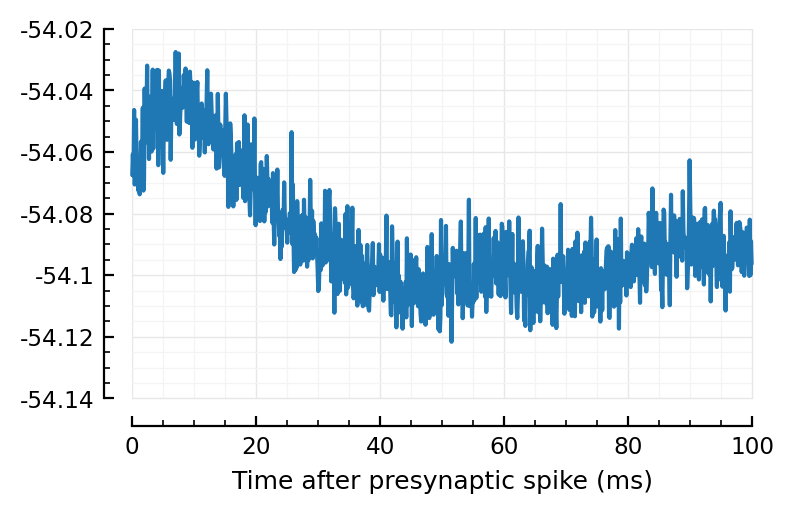
Wow much worse, with ceiled spikes.
So, obviously, let’s try and trim the spikes.
Also, what’s voltage histogram ey.
hist(sim_ceil.V / mV);
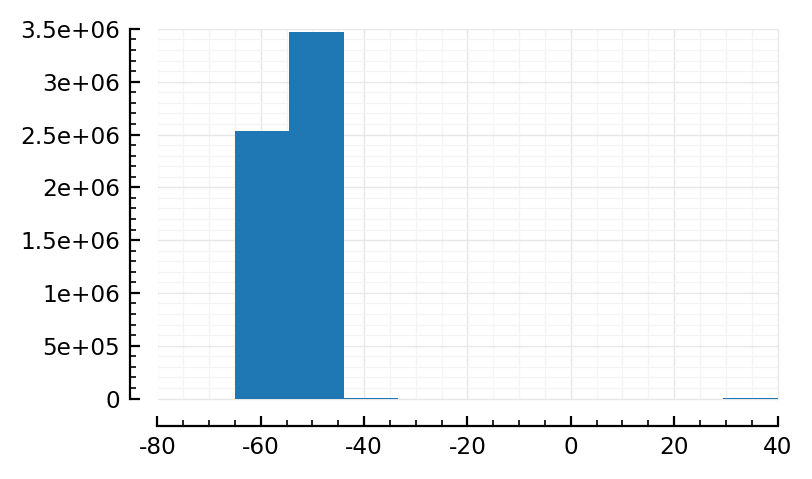
V = sim_ceil.V
hist(V[V .≥ -50mV] / mV);
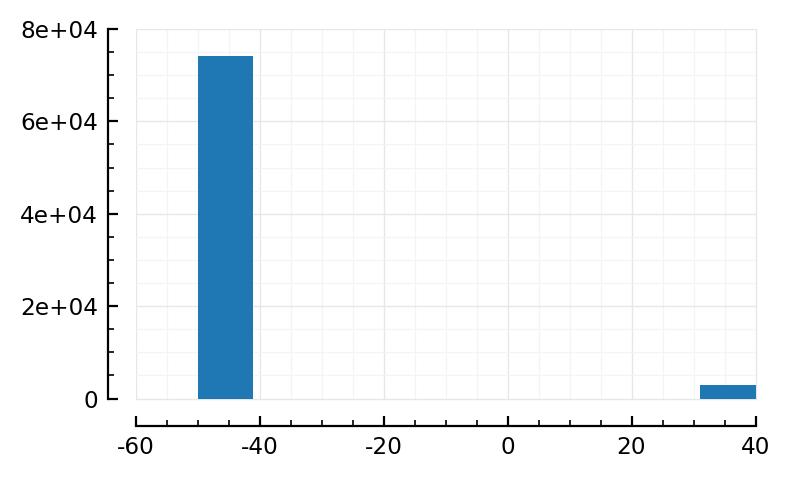
Can we do automatic selection? percentiles let’s see. Or outlier detection, or sth.
using Statistics
ps = [0, 0.01, 0.5, 0.9, 0.99, 0.999, 1] # i.e. 0, 1, 50, 90, 99, 99.1, 100-percentiles.
qs = quantile(V, ps) / mV
DataFrame("Proportion"=>ps, "Value (mV)"=>round.(qs, digits=1))
| Row | Proportion | Value (mV) |
|---|---|---|
| Float64 | Float64 | |
| 1 | 0.0 | -65.0 |
| 2 | 0.01 | -58.1 |
| 3 | 0.5 | -54.0 |
| 4 | 0.9 | -51.6 |
| 5 | 0.99 | -49.8 |
| 6 | 0.999 | -46.8 |
| 7 | 1.0 | 40.0 |
V_clip = copy(sim_ceil.V)
V_clip[V_clip .≥ -50mV] .= -50mV;
plotSTA(calc_STA(V_clip, exc_inputs[1].times));
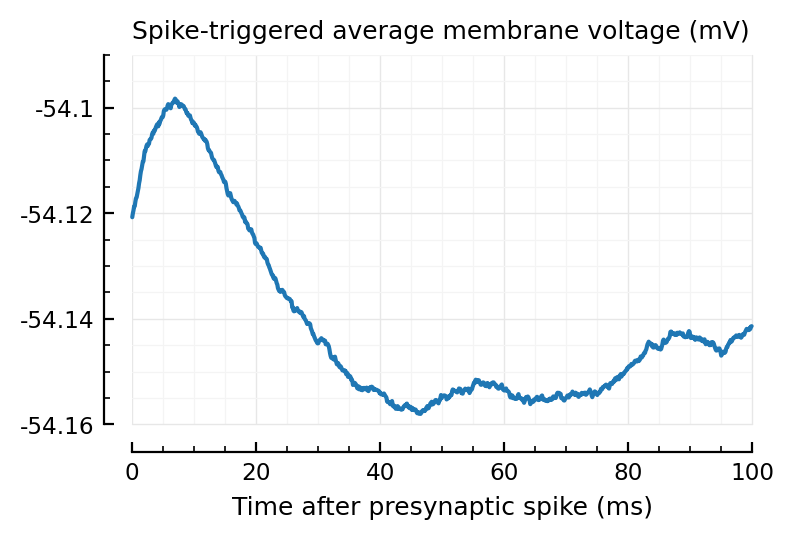
Holy damn :OOOO
This is so clean :OOOO.
plotSTA(calc_STA(V_clip, exc_inputs[100].times));
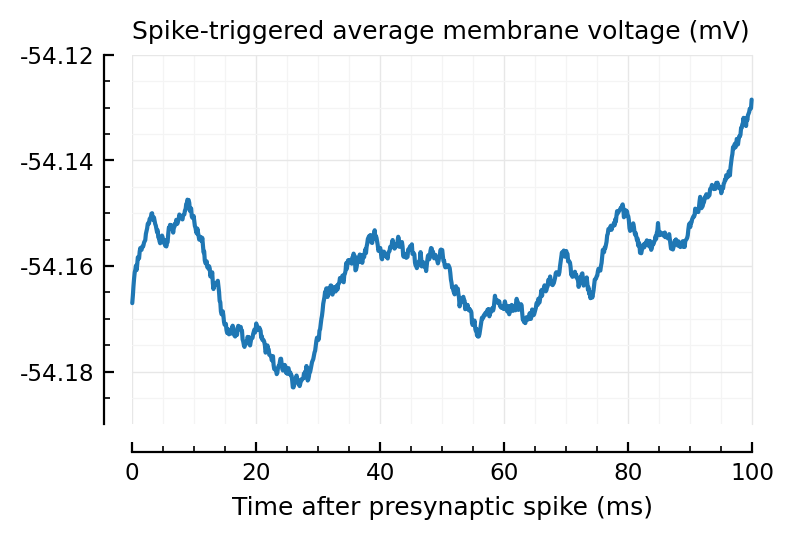
plotSTA(calc_STA(V_clip, exc_inputs[mid].times));
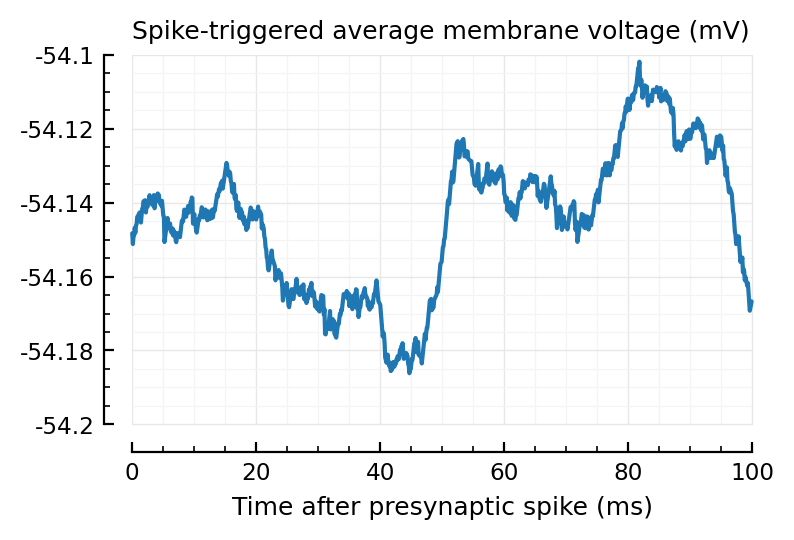
plotSTA(calc_STA(V_clip, exc_inputs[end].times));
Ok no real help for the lower spikers heh.
Sig itself¶
function ceilplot(; tlim, marker=nothing, ax, kw...)
plotsig(sim_ceil.V / mV, tlim, ms, label=".. with ceiled spikes"; ax, marker, kw...);
plotsig(sim.V / mV, tlim, ms, label="Original simulation"; ax, marker, kw...);
# legend(ax, reorder=[1=>2]);
end
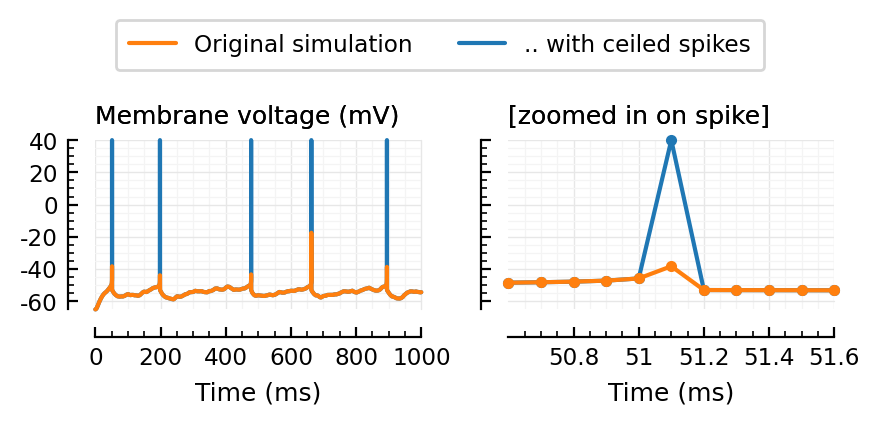
fig, axs = plt.subplots(ncols=2, figsize=(mtw, 0.4*mtw), sharey=true)
ceilplot(tlim = [0, 1000], ax=axs[0], hylabel="Membrane voltage (mV)");
ceilplot(tlim = [50.6, 51.6], marker=".", ax=axs[1], hylabel="[zoomed in on spike]");
l = axs[0].get_lines()
# rm_ticks_and_spine(axs[1], "left")
plt.figlegend(handles=[l[1], l[0]], ncols=2, loc="lower center", bbox_to_anchor=(0.5, 1))
plt.tight_layout();
‘What would V would be w/o thresholding’¶
@time sim_r = Nto1AdEx.sim(N, duration, record_all=true);
2.686393 seconds (6.03 M allocations: 1.383 GiB, 25.27% gc time)
We need to calc V. Which is Vprev + Δt * ΔV
t = sim_r.spiketimes[1]
t / ms
51.200000000000436
(; Δt, Eₑ, Eᵢ, Δₜ, Vₜ, gₗ, Eₗ, C) = Nto1AdEx
i = round(Int, t/Δt) # The spiketime `t` is one sample after where we want, but this i is correct
512
n = sim_r.recording[i];
(; V, gₑ, gᵢ, w) = n
V / mV
-38.24759825475466
n.DₜV
75.60870254277917
DₜV
966640.2938251279
Iₛ = gₑ*(V - Eₑ) + gᵢ*(V - Eᵢ)
DₜV = (-gₗ*(V - Eₗ) + gₗ*Δₜ*exp((V-Vₜ)/Δₜ) - Iₛ - w) / C
V_new = V + Δt * DₜV
V_new / mV
96625.78178425804
Heh.