2021-09-27 • Vary E/I proportion
Contents
2021-09-27 • Vary E/I proportion¶
Previously we just plotted STA’s of inhibitory inputs. Now we measure the STA height, and compare it to the STA height of shuffled input spike trains.
Then we calculate separate true positive rates for inhibitory and excitatory connections, and we do this for a range of p_inhibitory.
Prelude¶
from voltage_to_wiring_sim.notebook_init import *
Preloading: numpy, numba, matplotlib.pyplot, seaborn.
Importing from submodules … ✔
Imported `np`, `mpl`, `plt`, `sns`, `pd`
Imported codebase (`voltage_to_wiring_sim`) as `v`
Imported `*` from `v.support.units`
Setup autoreload
v.print_reproducibility_info()
This cell was last run by lpxtf3 on DUIP74576 on Thu 04 Nov 2021, at 16:30 (UTC+0000).
Last git commit (Wed 03 Nov 2021, 20:11).
Uncommited changes to 6 files.
Run simulation & connection tests¶
from voltage_to_wiring_sim.experiments.N_to_1_IE import Params, simulate_and_test_connections, indices_where
v.pprint(Params())
Params
------
sim_duration = 600
timestep = 0.0001
spike_rate = 20
Δg_syn = 8E-10
τ_syn = 0.007
neuron_params = {'C': 1e-10, 'a': 30.0, 'b': -2e-09, 'c': -0.05, ...}
imaging_spike_SNR = 20
v_syn_E = 0
v_syn_I = -0.07
num_spike_trains = 30
p_inhibitory = 0.6
p_connected = 0.6
window_duration = 0.1
rng_seed = 0
from voltage_to_wiring_sim.conntest.classification import apply_threshold
from voltage_to_wiring_sim.conntest.classification_IE import (
calc_AUCs,
evaluate_classification,
sweep_threshold,
)
@v.cache_to_disk("2021-09-16__vary_E_vs_I")
def sim_and_test_and_eval_performance(p: Params):
d, _, test_summaries = simulate_and_test_connections(p)
# Eval at fixed p_value threshold
is_classified_as_connected = apply_threshold(test_summaries, p_value_threshold=0.05)
evalu = evaluate_classification(
is_classified_as_connected, d.is_connected, d.is_inhibitory
)
# Eval at all p_value thresholds
thr_sweep = sweep_threshold(test_summaries, d.is_connected, d.is_inhibitory)
AUC_inh, AUC_exc = calc_AUCs(thr_sweep)
return (evalu.TPR_inh, evalu.TPR_exc, evalu.FPR, AUC_inh, AUC_exc)
from dataclasses import replace
from itertools import product
def set_reordered_legend(ax, order: tuple[int], **kwargs):
h, l = ax.get_legend_handles_labels()
ax.legend([h[i] for i in order], [l[i] for i in order], **kwargs)
color_inh = "C1"
color_exc = "C0"
color_unconn = "C2";
def vary_p_inhibited(base_params: Params, ax1, ax2):
p_inh = np.linspace(0, 1, num=11, endpoint=True)
seeds = [0, 1, 2, 3, 4, 5]
results = []
for p_i, seed in product(p_inh, seeds):
params = replace(base_params, p_inhibitory=p_i, rng_seed=seed)
results.append(sim_and_test_and_eval_performance(params))
M = np.reshape(results, (len(p_inh), len(seeds), -1))
TPR_inh, TPR_exc, FPR, AUC_inh, AUC_exc = (M[:,:,i] for i in range(M.shape[-1]))
ax1.plot(p_inh, TPR_inh, "o", c=color_inh, ms=6, alpha=0.3)
ax1.plot(p_inh, np.mean(TPR_inh, axis=1), "-", lw=5, label="Inhibitory conn.", c=color_inh)
ax1.plot(p_inh, FPR, "o", c=color_unconn, ms=5, alpha=0.3)
ax1.plot(p_inh, np.mean(FPR, axis=1), "-", lw=4, label="Non-connections", c=color_unconn)
ax1.plot(p_inh, TPR_exc, "o", c=color_exc, ms=4, alpha=0.3)
ax1.plot(p_inh, np.mean(TPR_exc, axis=1), "-", lw=2.6, label="Excitatory conn.", c=color_exc)
ax1.set_ylabel("Fraction detected")
set_reordered_legend(ax1, [2, 0, 1])
ax2.plot(p_inh, AUC_inh, "o", c=color_inh, ms=6, alpha=0.3)
ax2.plot(p_inh, AUC_exc, "o", c=color_exc, ms=4, alpha=0.3)
ax2.plot(p_inh, np.mean(AUC_inh, axis=1), "-", lw=5, label="Inhibitory conn.", c=color_inh)
ax2.plot(p_inh, np.mean(AUC_exc, axis=1), "-", lw=2.6, label="Excitatory conn.", c=color_exc)
ax2.set_ylim(0.5, 1.02)
ax2.set_xlabel("Proportion of inputs inhibitory ($p_{inh}$)")
ax2.set_ylabel("Area under ROC curve")
set_reordered_legend(ax2, [1, 0])
(An uncached run of the above takes a bit over 3 minutes, for 1 seed).
fig, axes = plt.subplots(2, 2, **v.figsize(width=1000), sharex=True)
vary_p_inhibited(Params(v_syn_I = -70 * mV), axes[0,0], axes[1,0])
vary_p_inhibited(Params(v_syn_I = -50 * mV), axes[0,1], axes[1,1])
set_reordered_legend(axes[0,1], [2,0,1], loc="center left", bbox_to_anchor=(0.02, 0.78))
axes[0,0].set_title("$v_{inh} = -70$ mV", pad=24)
axes[0,1].set_title("$v_{inh} = -50$ mV", pad=24)
plt.subplots_adjust(wspace=0.4)
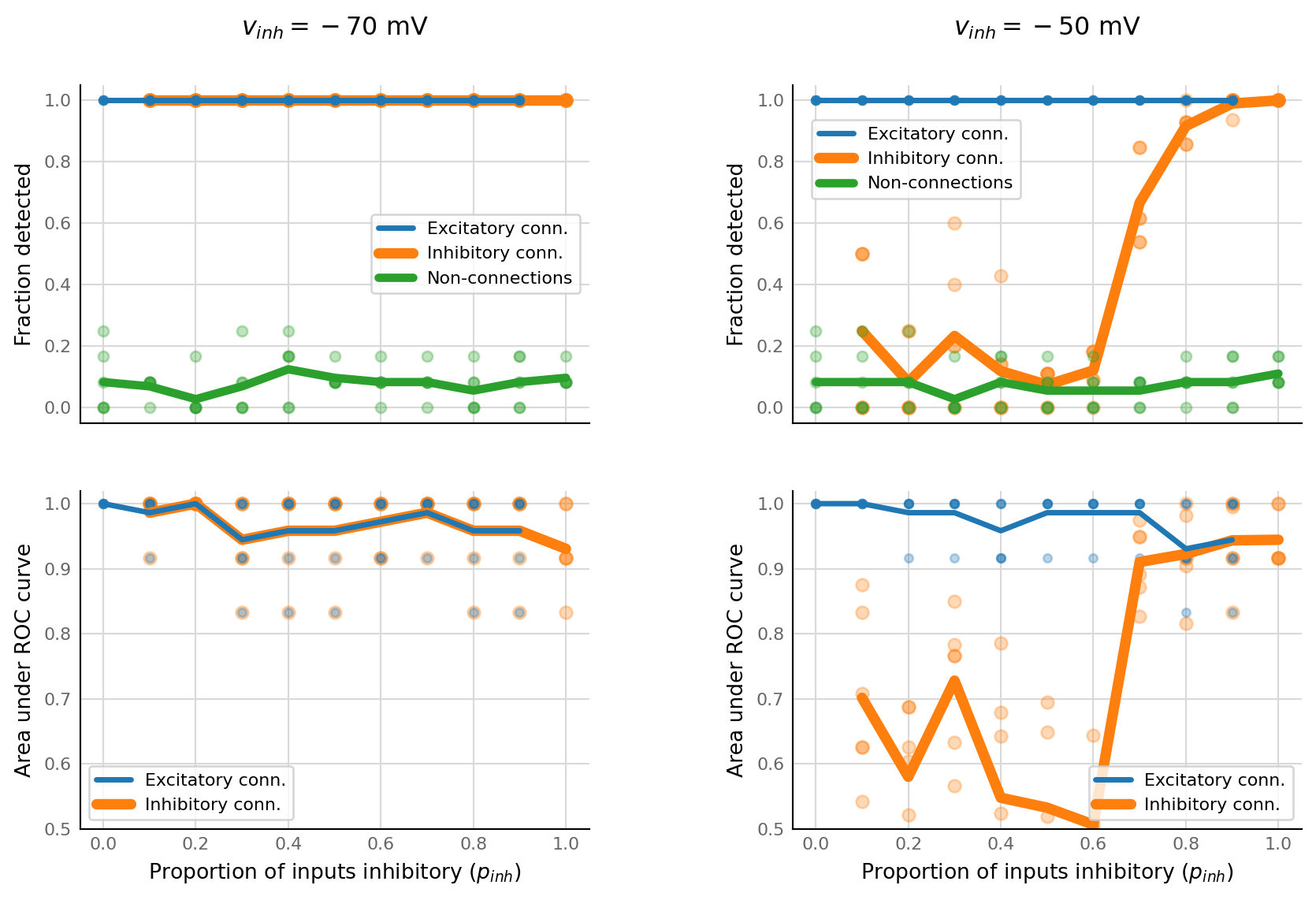
Uncached run for two seeds took 16’30”.
individual sims getting gradually slower.. from 20 s at start (v -70 p 0 seed 1) to 52 at end (v -50 p 1 seed 2). Maybe memory drag? python process is at 2GB now.
cache loading is deliciously fast though :)
Inspect some regimes¶
sim_and_test = v.cache_to_disk(simulate_and_test_connections);
fig, axes = plt.subplots(nrows=3, ncols=4, **v.figsize(width=1000))
for i, (v_syn_I, p_inh) in enumerate(product((-70 * mV, -50 * mV), (0.1, 0.9))):
d, td, ts = sim_and_test(Params(v_syn_I=v_syn_I, p_inhibitory=p_inh))
print(d.num_exc_conn, d.num_inh_conn)
ax = v.plot_signal(d.izh_output.V_m.slice(t_start=0, duration=1*second) / mV, time_units=ms, ax=axes[0, i])
ax.set_ylim(-65, -22);
ax.set_xlabel("Time (ms)")
ax.set_title("$v_{inh} = " + f"{v_syn_I / mV:.0f}$ mV" + "\n" +
"$p_{inh} = " + f"{p_inh:.1f}$", pad=24)
ax_e = v.plot_STA(td[indices_where(d.is_excitatory & d.is_connected)[0]].original_STA, ax=axes[1, i])
ax_i = v.plot_STA(td[indices_where(d.is_inhibitory & d.is_connected)[0]].original_STA, ax=axes[2, i])
ylims = (min(ax_i.get_ylim()[0], ax_e.get_ylim()[0]),
max(ax_i.get_ylim()[1], ax_e.get_ylim()[1]))
ax_e.set_ylim(ylims)
ax_i.set_ylim(ylims)
if i == 0:
ax.set_ylabel("Membrane pot. (mV)");
ax_e.set_ylabel("STA of VI signal for an" + '\n' + r"$\bf{excitatory}$ conn. (mV)")
ax_i.set_ylabel("STA of VI signal for an" + '\n' + r"$\bf{inhibitory}$ conn. (mV)")
else:
ax_e.set_ylabel(None)
ax_i.set_ylabel(None)
fig.tight_layout()
16 2
2 16
16 2
2 16
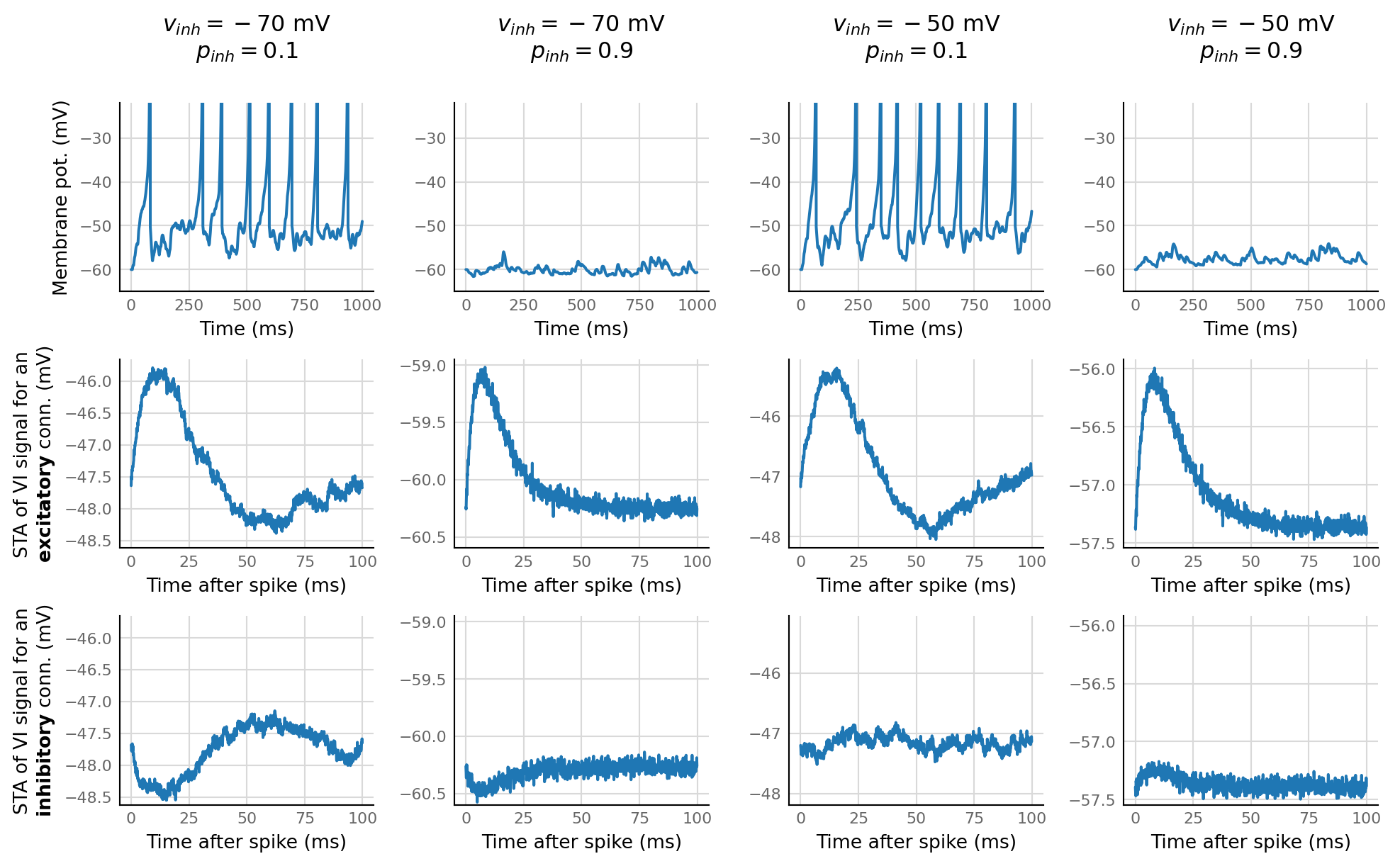
d, td, ts = sim_and_test(Params(v_syn_I=-50 * mV, p_inhibitory=0.1));
np.median(d.izh_output.V_m) / mV
-50.468785742174646
fig, axes = plt.subplots(nrows=3, ncols=4, **v.figsize(width=1000))
for i, (v_syn_I, p_inh) in enumerate(product((-50 * mV,), (0.5, 0.6, 0.7))):
d, td, ts = sim_and_test(Params(v_syn_I=v_syn_I, p_inhibitory=p_inh))
print(d.num_exc_conn, d.num_inh_conn)
ax = v.plot_signal(d.izh_output.V_m.slice(t_start=0, duration=8*second) / mV, time_units=ms, ax=axes[0, i])
ax.set_ylim(-65, -22);
ax.set_xlabel("Time (ms)")
ax.set_title("$v_{inh}$ = " + f"{v_syn_I / mV:.0f} mV" + "\n" +
"$p_{inh}$ = " + f"{p_inh:.1f}")
ax_e = v.plot_STA(td[indices_where(d.is_excitatory & d.is_connected)[0]].original_STA, ax=axes[1, i])
ax_i = v.plot_STA(td[indices_where(d.is_inhibitory & d.is_connected)[0]].original_STA, ax=axes[2, i])
ylims = (min(ax_i.get_ylim()[0], ax_e.get_ylim()[0]),
max(ax_i.get_ylim()[1], ax_e.get_ylim()[1]))
ax_e.set_ylim(ylims)
ax_i.set_ylim(ylims)
if i == 0:
ax.set_ylabel("Membrane pot. (mV)");
ax_e.set_ylabel("STA of VI signal for an" + '\n' + r"$\bf{excitatory}$ conn. (mV)")
ax_i.set_ylabel("STA of VI signal for an" + '\n' + r"$\bf{inhibitory}$ conn. (mV)")
else:
ax_e.set_ylabel(None)
ax_i.set_ylabel(None)
fig.tight_layout()
9 9
7 11
5 13
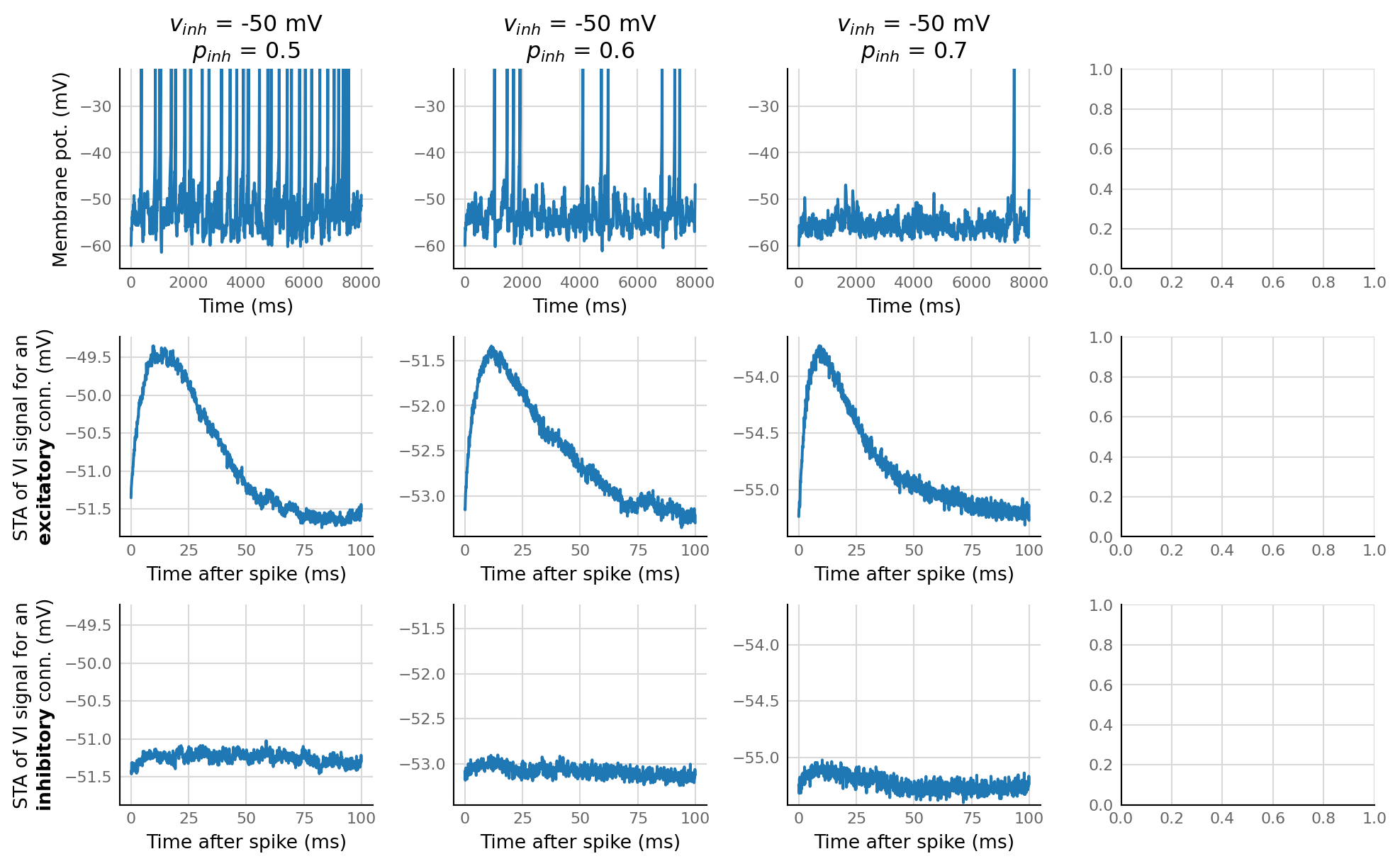
# v.plot_signal(d.VI_signal.slice(t_start=0, duration=1*second) / mV, time_units=ms);
Reproducibility¶
v.print_reproducibility_info(verbose=True)
This cell was last run by lpxtf3 on DUIP74576
on Thu 28 Oct 2021, at 15:09 (UTC+0100).
Last git commit (Thu 28 Oct 2021, 14:11).
Uncommited changes to:
M notebooks/2021-09-16__vary_E_vs_I.ipynb
Platform:
Windows-10
CPython 3.9.6 (C:\miniforge3\python.exe)
Intel(R) Xeon(R) W-2123 CPU @ 3.60GHz
Dependencies of voltage_to_wiring_sim and their installed versions:
numpy 1.21.1
matplotlib 3.4.2
numba 0.53.1
joblib 1.0.1
seaborn 0.11.1
scipy 1.7.0
preload 2.2
nptyping 1.4.2
Full conda list:
# packages in environment at C:\miniforge3:
#
# Name Version Build Channel
argon2-cffi 20.1.0 py39hb82d6ee_2 conda-forge
async_generator 1.10 py_0 conda-forge
attrs 21.2.0 pyhd8ed1ab_0 conda-forge
backcall 0.2.0 pyh9f0ad1d_0 conda-forge
backports 1.0 py_2 conda-forge
backports.functools_lru_cache 1.6.4 pyhd8ed1ab_0 conda-forge
bleach 3.3.1 pyhd8ed1ab_0 conda-forge
brotlipy 0.7.0 py39hb82d6ee_1001 conda-forge
ca-certificates 2019.11.28 hecc5488_0 conda-forge/label/cf202003
certifi 2021.10.8 py39hcbf5309_0 conda-forge
cffi 1.14.6 py39h0878f49_0 conda-forge
chardet 4.0.0 py39hcbf5309_1 conda-forge
charset-normalizer 2.0.0 pyhd8ed1ab_0 conda-forge
click 7.1.2 pypi_0 pypi
colorama 0.4.4 pyh9f0ad1d_0 conda-forge
conda 4.10.3 py39hcbf5309_2 conda-forge
conda-package-handling 1.7.3 py39hb3671d1_0 conda-forge
cryptography 3.4.7 py39hd8d06c1_0 conda-forge
cycler 0.10.0 pypi_0 pypi
debugpy 1.4.1 py39h415ef7b_0 conda-forge
decorator 5.0.9 pyhd8ed1ab_0 conda-forge
defusedxml 0.7.1 pyhd8ed1ab_0 conda-forge
entrypoints 0.3 pyhd8ed1ab_1003 conda-forge
icu 68.2 h0e60522_0 conda-forge
idna 3.1 pyhd3deb0d_0 conda-forge
importlib-metadata 4.6.1 py39hcbf5309_0 conda-forge
ipykernel 6.0.3 py39h832f523_0 conda-forge
ipython 7.25.0 py39h832f523_1 conda-forge
ipython_genutils 0.2.0 py_1 conda-forge
jedi 0.18.0 py39hcbf5309_2 conda-forge
jinja2 3.0.1 pyhd8ed1ab_0 conda-forge
joblib 1.0.1 pypi_0 pypi
jpeg 9d h8ffe710_0 conda-forge
jsonschema 3.2.0 pyhd8ed1ab_3 conda-forge
jupyter_client 6.1.12 pyhd8ed1ab_0 conda-forge
jupyter_contrib_core 0.3.3 py_2 conda-forge
jupyter_contrib_nbextensions 0.5.1 pyhd8ed1ab_2 conda-forge
jupyter_core 4.7.1 py39hcbf5309_0 conda-forge
jupyter_highlight_selected_word 0.2.0 py39hcbf5309_1002 conda-forge
jupyter_latex_envs 1.4.6 pyhd8ed1ab_1002 conda-forge
jupyter_nbextensions_configurator 0.4.1 py39hcbf5309_2 conda-forge
jupyterlab_pygments 0.1.2 pyh9f0ad1d_0 conda-forge
jupytray 0.7 pypi_0 pypi
kiwisolver 1.3.1 pypi_0 pypi
libclang 11.1.0 default_h5c34c98_1 conda-forge
libiconv 1.16 he774522_0 conda-forge
libpng 1.6.37 h1d00b33_2 conda-forge
libsodium 1.0.18 h8d14728_1 conda-forge
libxml2 2.9.12 hf5bbc77_0 conda-forge
libxslt 1.1.33 h65864e5_2 conda-forge
libzlib 1.2.11 h8ffe710_1013 conda-forge
llvmlite 0.36.0 pypi_0 pypi
lxml 4.6.3 py39h4fd7cdf_0 conda-forge
markupsafe 2.0.1 py39hb82d6ee_0 conda-forge
matplotlib 3.4.2 pypi_0 pypi
matplotlib-inline 0.1.2 pyhd8ed1ab_2 conda-forge
menuinst 1.4.17 py39hcbf5309_1 conda-forge
miniforge_console_shortcut 2.0 h57928b3_0 conda-forge
mistune 0.8.4 py39hb82d6ee_1004 conda-forge
nbclient 0.5.3 pyhd8ed1ab_0 conda-forge
nbconvert 6.1.0 py39hcbf5309_0 conda-forge
nbformat 5.1.3 pyhd8ed1ab_0 conda-forge
nest-asyncio 1.5.1 pyhd8ed1ab_0 conda-forge
notebook 6.4.0 pyha770c72_0 conda-forge
nptyping 1.4.2 pypi_0 pypi
numba 0.53.1 pypi_0 pypi
numpy 1.21.1 pypi_0 pypi
openssl 1.1.1l h8ffe710_0 conda-forge
packaging 21.0 pyhd8ed1ab_0 conda-forge
pandas 1.3.1 pypi_0 pypi
pandoc 2.14.1 h8ffe710_0 conda-forge
pandocfilters 1.4.2 py_1 conda-forge
parso 0.8.2 pyhd8ed1ab_0 conda-forge
pickleshare 0.7.5 py_1003 conda-forge
pillow 8.3.1 pypi_0 pypi
pip 21.2.1 pyhd8ed1ab_0 conda-forge
preload 2.2 pypi_0 pypi
prometheus_client 0.11.0 pyhd8ed1ab_0 conda-forge
prompt-toolkit 3.0.19 pyha770c72_0 conda-forge
pycosat 0.6.3 py39hb82d6ee_1006 conda-forge
pycparser 2.20 pyh9f0ad1d_2 conda-forge
pygments 2.9.0 pyhd8ed1ab_0 conda-forge
pyopenssl 20.0.1 pyhd8ed1ab_0 conda-forge
pyparsing 2.4.7 pyh9f0ad1d_0 conda-forge
pyqt 5.12.3 py39hcbf5309_7 conda-forge
pyqt-impl 5.12.3 py39h415ef7b_7 conda-forge
pyqt5-sip 4.19.18 py39h415ef7b_7 conda-forge
pyqtchart 5.12 py39h415ef7b_7 conda-forge
pyqtwebengine 5.12.1 py39h415ef7b_7 conda-forge
pyrsistent 0.17.3 py39hb82d6ee_2 conda-forge
pysocks 1.7.1 py39hcbf5309_3 conda-forge
python 3.9.6 h7840368_1_cpython conda-forge
python-dateutil 2.8.2 pyhd8ed1ab_0 conda-forge
python_abi 3.9 2_cp39 conda-forge
pytz 2021.1 pypi_0 pypi
pywin32 300 py39hb82d6ee_0 conda-forge
pywinpty 1.1.3 py39h99910a6_0 conda-forge
pyyaml 6.0 py39hb82d6ee_0 conda-forge
pyzmq 22.1.0 py39he46f08e_0 conda-forge
qt 5.12.9 h5909a2a_4 conda-forge
requests 2.26.0 pyhd8ed1ab_0 conda-forge
ruamel_yaml 0.15.80 py39hb82d6ee_1004 conda-forge
scipy 1.7.0 pypi_0 pypi
seaborn 0.11.1 pypi_0 pypi
send2trash 1.7.1 pyhd8ed1ab_0 conda-forge
setuptools 49.6.0 py39hcbf5309_3 conda-forge
six 1.16.0 pyh6c4a22f_0 conda-forge
sqlite 3.36.0 h8ffe710_0 conda-forge
terminado 0.10.1 py39hcbf5309_0 conda-forge
testpath 0.5.0 pyhd8ed1ab_0 conda-forge
tornado 6.1 py39hb82d6ee_1 conda-forge
tqdm 4.61.2 pyhd8ed1ab_1 conda-forge
traitlets 5.0.5 py_0 conda-forge
typish 1.9.2 pypi_0 pypi
tzdata 2021a he74cb21_1 conda-forge
ucrt 10.0.20348.0 h57928b3_0 conda-forge
urllib3 1.26.6 pyhd8ed1ab_0 conda-forge
vc 14.2 hb210afc_5 conda-forge
voltage-to-wiring-sim 0.1 dev_0 <develop>
vs2015_runtime 14.29.30037 h902a5da_5 conda-forge
wcwidth 0.2.5 pyh9f0ad1d_2 conda-forge
webencodings 0.5.1 py_1 conda-forge
wheel 0.36.2 pyhd3deb0d_0 conda-forge
win_inet_pton 1.1.0 py39hcbf5309_2 conda-forge
wincertstore 0.2 py39hcbf5309_1006 conda-forge
winpty 0.4.3 4 conda-forge
winshell 0.6 pypi_0 pypi
yaml 0.2.5 he774522_0 conda-forge
zeromq 4.3.4 h0e60522_0 conda-forge
zipp 3.5.0 pyhd8ed1ab_0 conda-forge
zlib 1.2.11 h8ffe710_1013 conda-forge