2022-07-14 • Not directly connected but detected – with self-bug
Contents
2022-07-14 • Not directly connected but detected – with self-bug¶
Imports¶
#
using Revise
using MyToolbox
using VoltoMapSim
[ Info: Precompiling VoltoMapSim [f713100b-c48c-421a-b480-5fcb4c589a9e]
Params¶
Based on Roxin (see previous nb).
d = 6
p = get_params(
duration = 10minutes,
p_conn = 0.04,
g_EE = 1 / d,
g_EI = 18 / d,
g_IE = 36 / d,
g_II = 31 / d,
ext_current = Normal(-0.5 * pA/√seconds, 5 * pA/√seconds),
E_inh = -80 * mV,
to_record = [1, 801],
);
# dumps(p)
Conntest¶
m = 1 # ID of recorded excitatory neuron
v = s.signals[m].v
ii = get_input_info(m, s, p);
ii.num_inputs
(exc = 26, inh = 10)
length(ii.unconnected_neurons)
964
perf = evaluate_conntest_perf(v, ii.spiketrains, p);
Testing connections: 100%|██████████████████████████████| Time: 0:00:29
perf.detection_rates
(TPR_exc = 0.154, TPR_inh = 1, FPR = 0.15)
So we’re investigating those 15% false positives, which would be 5% if directly-unconnected spiketrains were not related to the voltage signal.
signif_unconn = findall(perf.p_values.unconn .< p.evaluation.α)
# These are indices in `ii.unconnected_neurons`
6-element Vector{Int64}:
1
3
4
15
21
35
Note that these are indices in unconnected_neurons, not global neuron IDs.
length(signif_unconn) / p.evaluation.N_tested_presyn
0.15
(The eval_conntest_perf function takes the first N_tested_presyn = 40 of the spiketrains it’s given).
STAs¶
for i in signif_unconn
_, ax = plt.subplots(figsize=(2.2, 1.8))
plotSTA(v, ii.spiketrains.unconn[i], p; ax, ylim = (-60.2, -51))
end
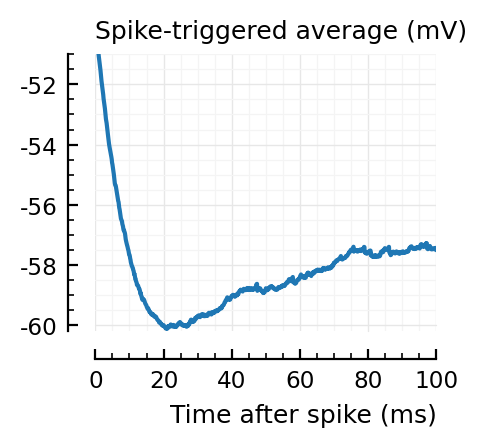
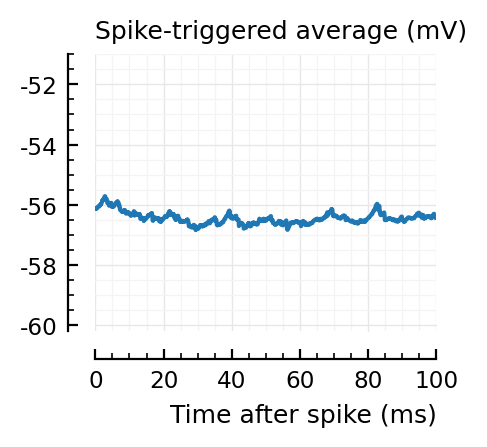
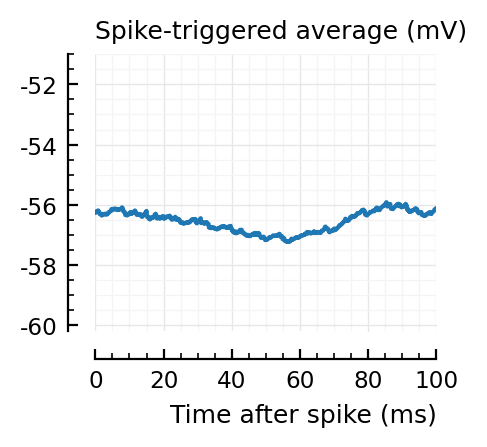
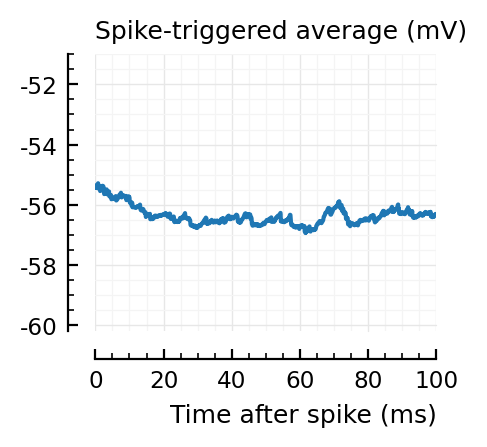
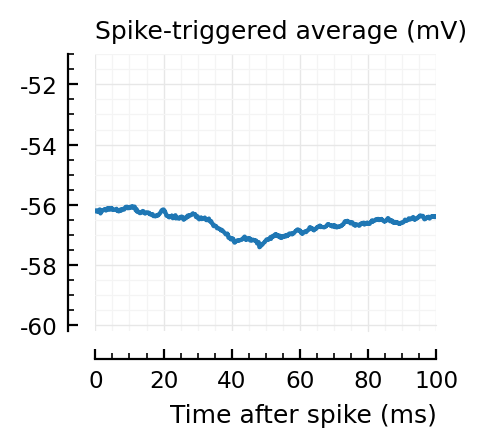
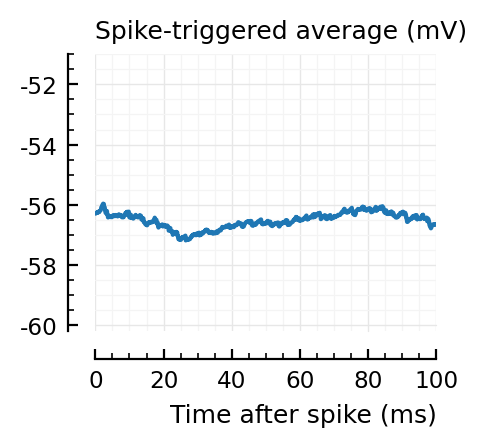
A bug: the first “unconnected” one (1) is the neuron itself.
Fixed self bug¶
Editing input_info and redoing the above.
ii = get_input_info(m, s, p);
(Should have given this another name and not overwritten the previous)
length(ii.unconnected_neurons)
963
perf = evaluate_conntest_perf(v, ii.spiketrains, p);
Testing connections: 100%|██████████████████████████████| Time: 0:01:00m
perf.detection_rates
(TPR_exc = 0.154, TPR_inh = 1, FPR = 0.15)
signif_unconn = ii.unconnected_neurons[findall(perf.p_values.unconn .< p.evaluation.α)]
6-element Vector{Int64}:
4
5
14
16
22
37
These are now global neuron IDs – less confusing to work with.
STAs = [calc_STA(v, s.spike_times[n], p) for n in signif_unconn]
ylim = [minimum([minimum(S) for S in STAs]), maximum([maximum(S) for S in STAs])] ./ mV
for n in signif_unconn
_, ax = plt.subplots(figsize=(2.2, 1.8))
plotSTA(v, s.spike_times[n], p; ax, ylim)
end
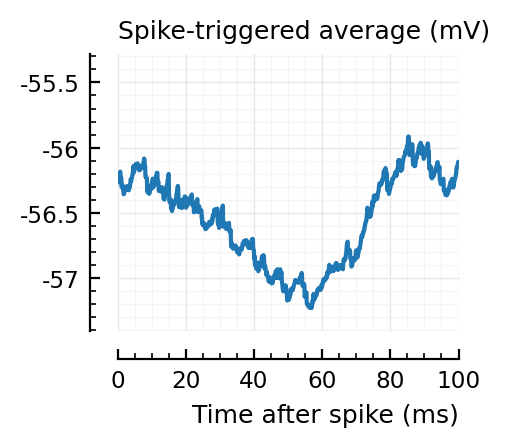
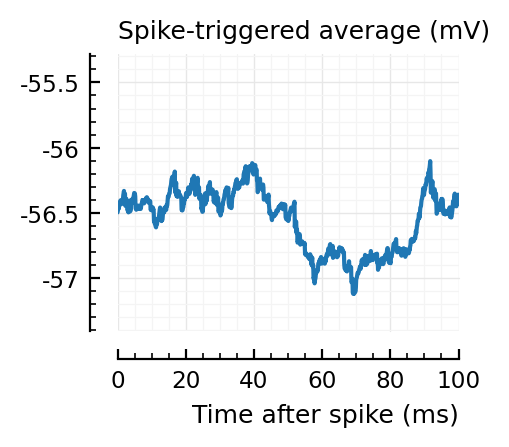
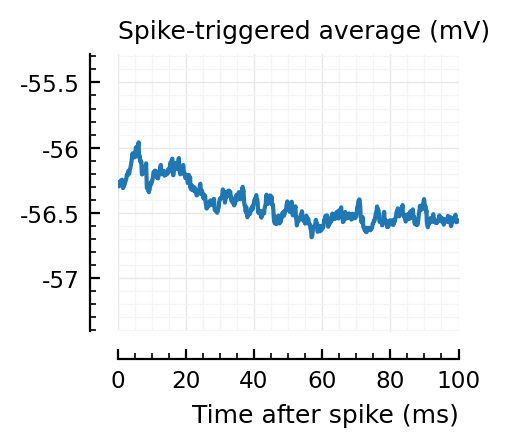
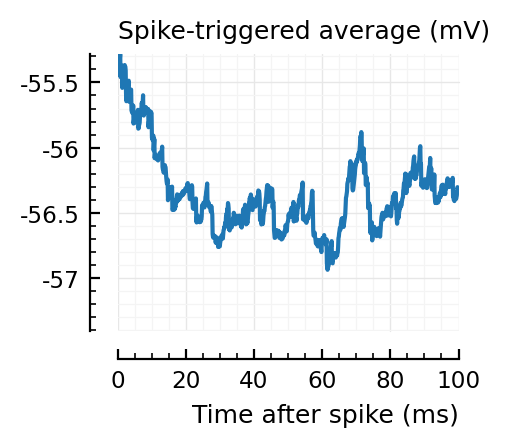
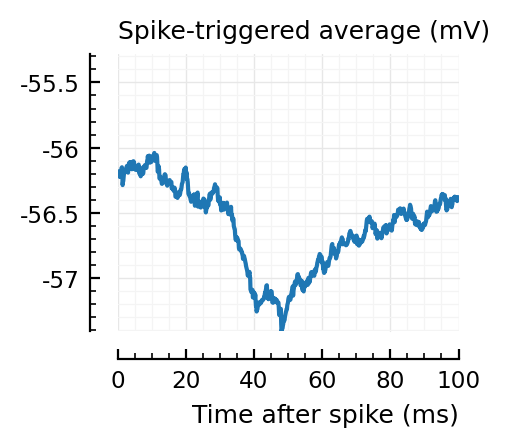
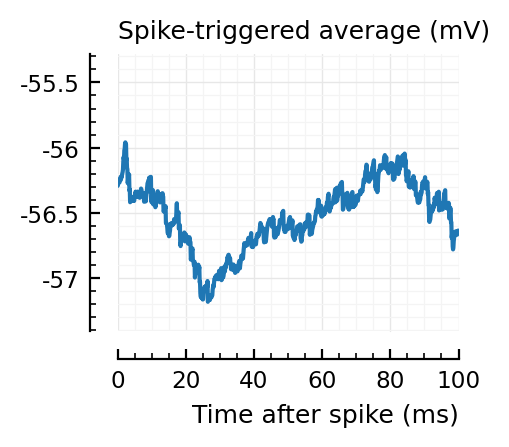
We expect 2/6 (so 2 / 40 tested, i.e. 5%) to change, depending on shuffle.
So let’s try another test.
We do need to set the rng seed manually as the test sets it for reproducibility.
p2 = @set p.evaluation.rngseed = 1;
perf2 = evaluate_conntest_perf(v, ii.spiketrains, p2);
Testing connections: 100%|██████████████████████████████| Time: 0:01:04
perf2.detection_rates
(TPR_exc = 0.115, TPR_inh = 1, FPR = 0.125)
signif_unconn2 = ii.unconnected_neurons[findall(perf2.p_values.unconn .< p.evaluation.α)]
5-element Vector{Int64}:
4
14
16
22
37
signif_unconn
6-element Vector{Int64}:
4
5
14
16
22
37
So all except 5 are common between both shuffle seeds.